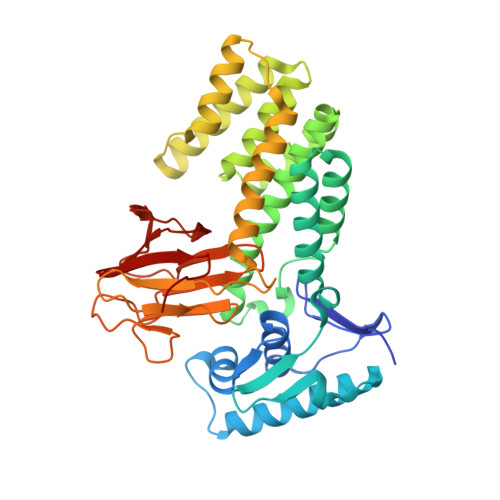
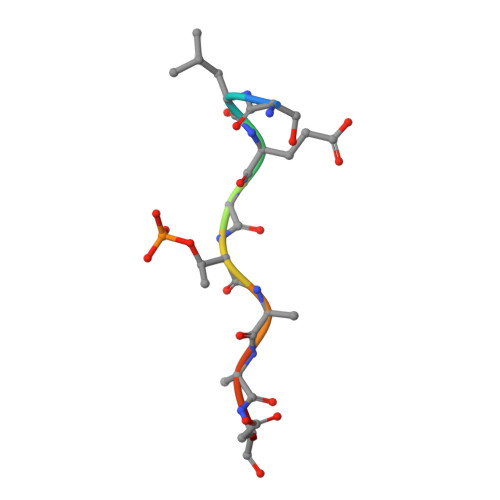
Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis.
Alderwick, L.J., Molle, V., Kremer, L., Cozzone, A.J., Dafforn, T.R., Besra, G.S., Futterer, K.(2006) Proc Natl Acad Sci U S A 103: 2558-2563
- PubMed: 16477027
- DOI: https://doi.org/10.1073/pnas.0507766103
- Primary Citation of Related Structures:
2FEZ, 2FF4 - PubMed Abstract:
Ser/Thr phosphorylation has emerged as a critical regulatory mechanism in a number of bacteria, including Mycobacterium tuberculosis. This problematic pathogen encodes 11 eukaryotic-like Ser/Thr kinases, yet few substrates or signaling targets have been characterized. Here, we report the structure of EmbR (2.0 A), a putative transcriptional regulator of key arabinosyltransferases (EmbC, -A, and -B), and an endogenous substrate of the Ser/Thr-kinase PknH. EmbR presents a unique domain architecture: the N-terminal winged-helix DNA-binding domain forms an extensive interface with the all-helical central bacterial transcriptional activation domain and is positioned adjacent to the regulatory C-terminal forkhead-associated (FHA) domain, which mediates binding to a Thr-phosphorylated site in PknH. The structure in complex with a phospho-peptide (1.9 A) reveals a conserved mode of phospho-threonine recognition by the FHA domain and evidence for specific recognition of the cognate kinase. The present structures suggest hypotheses as to how EmbR might propagate the phospho-relay signal from its cognate kinase, while serving as a template for the structurally uncharacterized Streptomyces antibiotic regulatory protein family of transcription factors.
Organizational Affiliation:
School of Biosciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom.