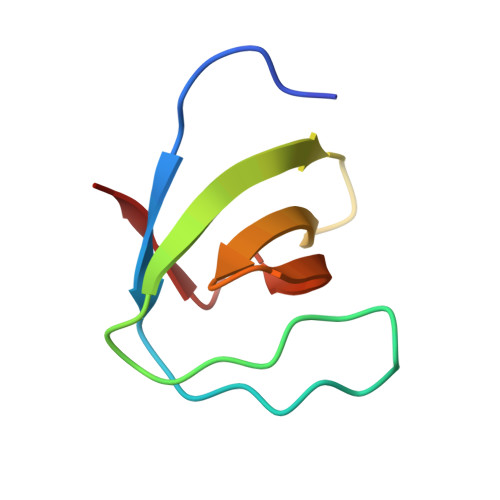
Crystal structure of the N-terminal SH3 domain of mouse betaPIX, p21-activated kinase-interacting exchange factor
Li, X., Liu, X., Sun, F., Gao, J., Zhou, H., Gao, G.F., Bartlam, M., Rao, Z.(2006) Biochem Biophys Res Commun 339: 407-414
- PubMed: 16307729
- DOI: https://doi.org/10.1016/j.bbrc.2005.10.212
- Primary Citation of Related Structures:
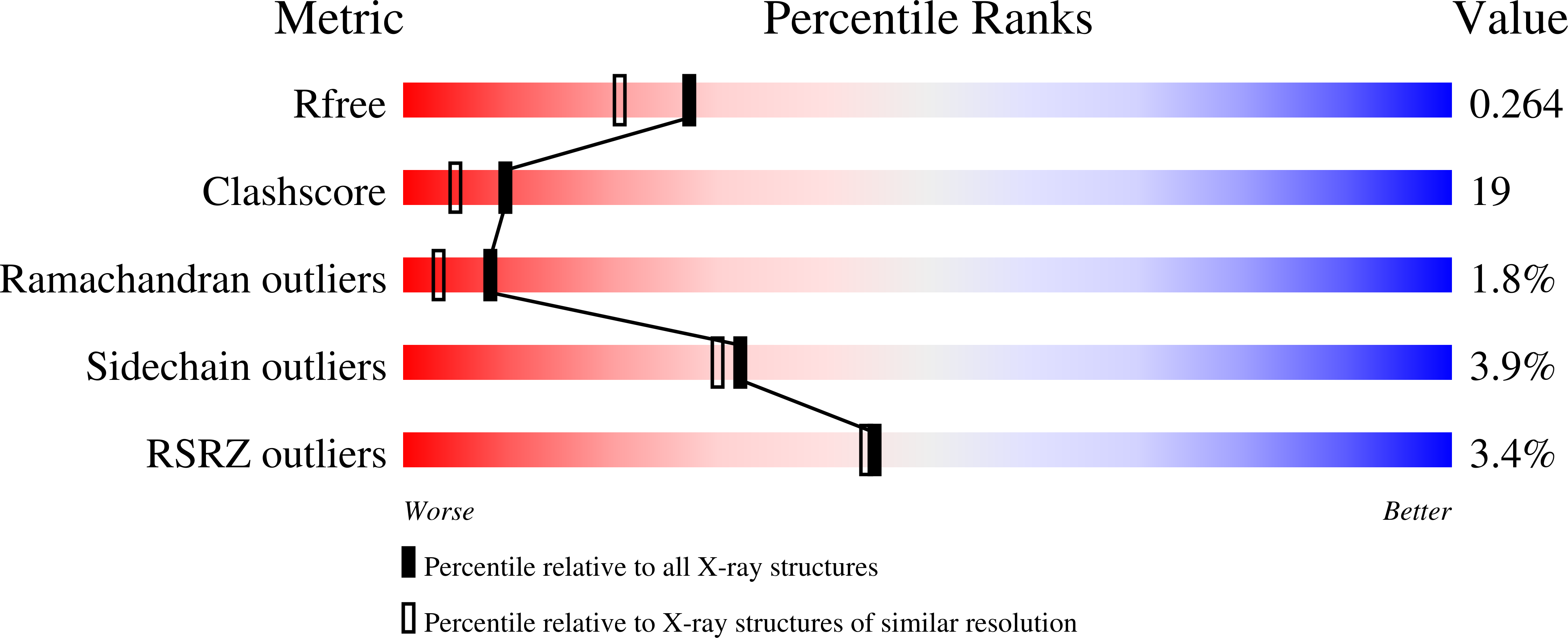
2ESW - PubMed Abstract:
The mouse betaPIX-SH3 domain, residues 8-63 of P21-activated kinase interacting exchange factor, has been characterized by X-ray diffraction. Crystals belonging to space group P3(2)21 diffracted to 2.0 A and the structure was phased by the single-wavelength anomalous diffraction method. The domain is a compact beta-barrel with an overall conformation similar to the general SH3 structure. The X-ray structure shows mouse betaPIX-SH3 domain binding the way in which the betaPIX characteristic amino acids do so for an unconventional ligand binding surface. This arrangement provides a rationale for the unusual ligand recognition motif exhibited by mouse betaPIX-SH3 domain. Comparison with another SH3/peptide complex shows that the recognition mode of the mouse betaPIX-SH3 domain should be very similar to the RXXK ligand binding mode. The unique large and planar hydrophobic pocket may contribute to the promiscuity of betaPIX-SH3 domain resulting in its multiple biological functions.
Organizational Affiliation:
Laboratory of Structural Biology, Tsinghua University, Beijing 100084, China.