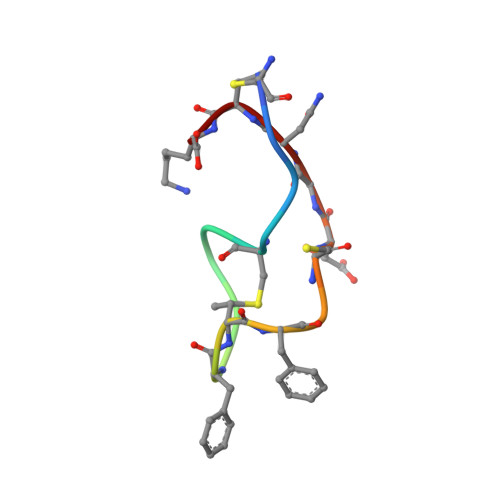
Structure determination of an immunopotentiator peptide, cinnamycin, complexed with lysophosphatidylethanolamine by 1H-NMR1.
Hosoda, K., Ohya, M., Kohno, T., Maeda, T., Endo, S., Wakamatsu, K.(1996) J Biochem 119: 226-230
- PubMed: 8882709
- DOI: https://doi.org/10.1093/oxfordjournals.jbchem.a021226
- PubMed Abstract:
The three-dimensional structure of a complex of cinnamycin, a 19-amino acid residue immunopotentiator peptide, and lysophosphatidylethanolamine was determined by 1H-NMR. The complex was cylindrical in shape, 11 A in diameter and 26 A in length, excluding the acyl chain of the phospholipid. The peptide had a hydrophobic pocket surrounded by residues Phe-7 through Ala(S)-14 to bind to the head group of the ligand. Fitting of the head group to the hydrophobic pocket was so good that other than a glycerophosphoethanolamine head group would be unable to fit the pocket. The goodness of the fitting is compatible with the strict specificity of ligand binding of the peptide.
Organizational Affiliation:
Department of Biochemical Sciences, Gunma University.