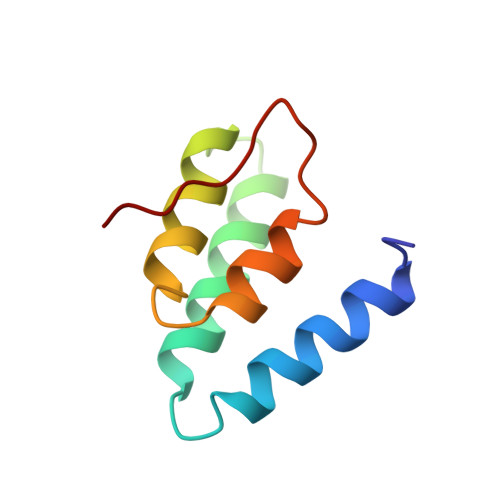
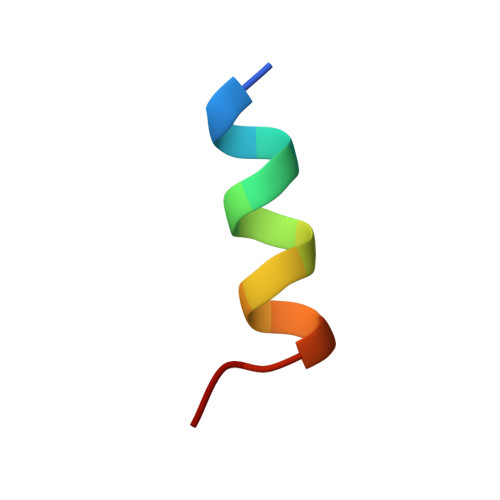
The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
Nomura, M., Uda-Tochio, H., Murai, K., Mori, N., Nishimura, Y.(2005) J Mol Biol 354: 903-915
- PubMed: 16288918
- DOI: https://doi.org/10.1016/j.jmb.2005.10.008
- Primary Citation of Related Structures:
2CZY - PubMed Abstract:
In non-neuronal cells and neuronal progenitors, many neuron-specific genes are repressed by a neural restrictive silencer factor (NRSF)/repressor element 1 silencing transcription factor (REST), which is an essential transcriptional repressor recruiting the Sin3-HDAC complex. Sin3 contains four paired amphipathic helix (PAH) domains, PAH1, PAH2, PAH3 and PAH4. A specific target repressor for Sin3 is likely to bind to one of them independently. So far, only the tertiary structures of PAH2 domain complexes, when bound to the Sin3-interacting domains of Mad1 and HBP1, have been determined. Here, we reveal that the N-terminal repressor domain of NRSF/REST binds to the PAH1 domain of mSin3B, and determine the structure of the PAH1 domain associated with the NRSF/REST minimal repressor domain. Compared to the PAH2 structure, PAH1 holds a rather globular four-helix bundle structure with a semi-ordered C-terminal tail. In contrast to the amphipathic alpha-helix of Mad1 or HBP1 bound to PAH2, the short hydrophobic alpha-helix of NRSF/REST is captured in the cleft of PAH1. A nuclear hormone receptor corepressor, N-CoR has been found to bind to the PAH1 domain with a lower affinity than NRSF/REST by using its C-terminal region, which contains fewer hydrophobic amino acid residues than the NRSF/REST helix. For strong binding to a repressor, PAH1 seems to require a short alpha-helix consisting of mostly hydrophobic amino acid residues within the repressor. Each of the four PAH domains of Sin3 seems to interact with a characteristic helix of a specific repressor; PAH1 needs a mostly hydrophobic helix and PAH2 needs an amphipathic helix in each target repressor.
Organizational Affiliation:
Graduate School of Supramolecular Biology Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.