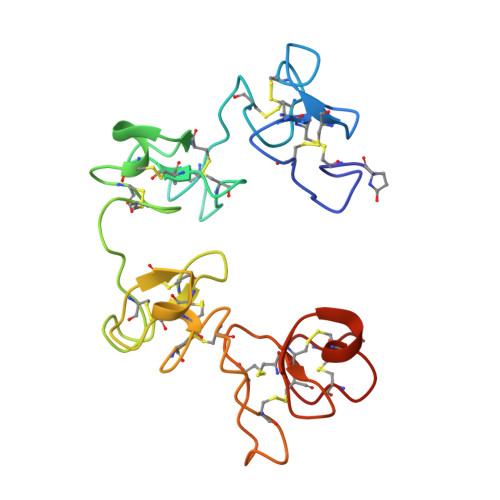
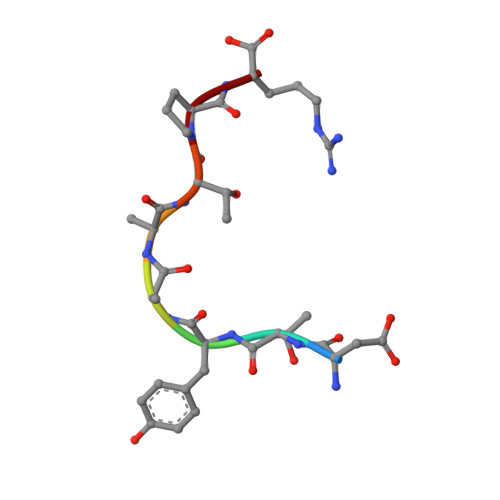
Crystallographic refinement and structure analysis of the complex of wheat germ agglutinin with a bivalent sialoglycopeptide from glycophorin A.
Wright, C.S., Jaeger, J.(1993) J Mol Biol 232: 620-638
- PubMed: 8345526
- DOI: https://doi.org/10.1006/jmbi.1993.1415
- Primary Citation of Related Structures:
2CWG - PubMed Abstract:
Wheat germ agglutinin (WGA) elicits a number of biological effects in erythrocytes as a result of specific binding to the transmembrane protein glycophorin A. The structure of co-crystals of WGA (isolectin 1: WGA1) with a bivalent sialoglycopeptide fragment of glycophorin A (T5), determined at 2.0 A resolution, has been further refined and analyzed with respect to ligand-induced changes in the tertiary structure, mobility, solvation, saccharide conformation and protein/saccharide interactions at three independent N-acetyl-D-neuraminic (NeuNAc) binding sites. The final model, which includes the two independent WGA1 monomers (composed of domains A, B, C and D), two positions for bound T5 sialo-tetrasaccharide (NeuNAc-alpha 2,3-Gal-beta 1,3-(alpha 2,6-NeuNAc)GalNAc) and 386 water molecules, refined to a crystallographic R-factor of 17.1% (Fo > 1.0 sigma) and an average temperature factor of 31.99 A2. Comparisons between the tertiary structures of the liganded and unliganded WGA1 dimers indicate that the largest deviations from 2-fold symmetry are localized in domains engaged in sugar binding (B1 and C2) and at the C-terminal domain of monomer II (D2), forming a strong lattice contact. Interactions of the tetrasaccharide with amino acid ligands in the three binding sites and with water were carefully analyzed and compared. Bound conformations of terminal NeuNAc match to within a root-mean-square delta r of 0.3 A. The specificity-determining N-acetyl group superimposes best in comparison with other substituents of the sugar ring. Of the five domain binding sites that are not occupied in this dimeric crosslinked complex, only one is accessible to the NeuNAc monosaccharide as determined from a difference Fourier map at 3.0 A resolution.
Organizational Affiliation:
Department of Biochemistry, Medical College of Virginia, Virginia Commonwealth University, Richmond 23298.