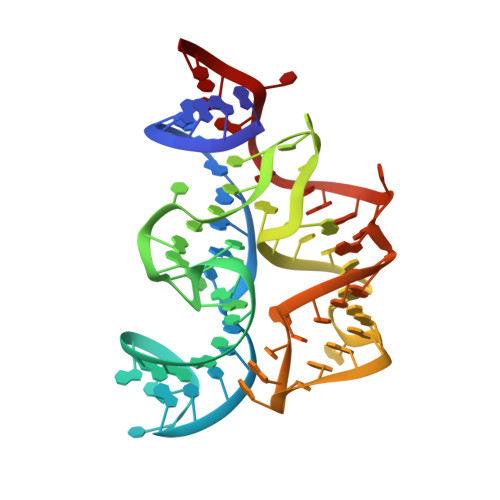
Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Thore, S., Leibundgut, M., Ban, N.(2006) Science 312: 1208
- PubMed: 16675665
- DOI: https://doi.org/10.1126/science.1128451
- Primary Citation of Related Structures:
2CKY - PubMed Abstract:
Riboswitches are untranslated regions of messenger RNA, which adopt alternate structures depending on the binding of specific metabolites. Such conformational switching regulates the expression of proteins involved in the biosynthesis of riboswitch substrates. Here, we present the 2.9 angstrom-resolution crystal structure of the eukaryotic Arabidopsis thaliana thiamine pyrophosphate (TPP)-specific riboswitch in complex with its natural ligand. The riboswitch specifically recognizes the TPP via conserved residues located within two highly distorted parallel "sensor" helices. The structure provides the basis for understanding the reorganization of the riboswitch fold upon TPP binding and explains the mechanism of resistance to the antibiotic pyrithiamine.
Organizational Affiliation:
ETH Zurich, Institute of Molecular Biology and Biophysics, 8092 Zurich, Switzerland. ban@mol.biol.ethz.ch