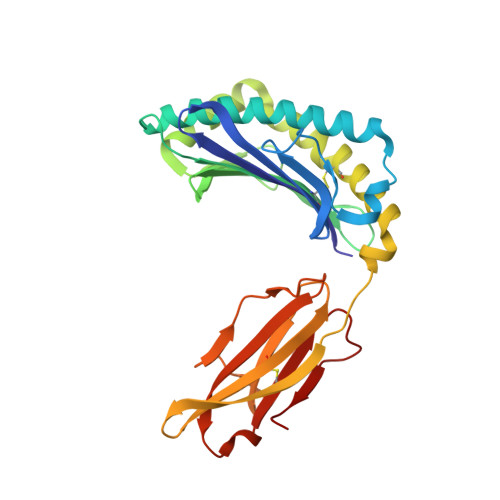
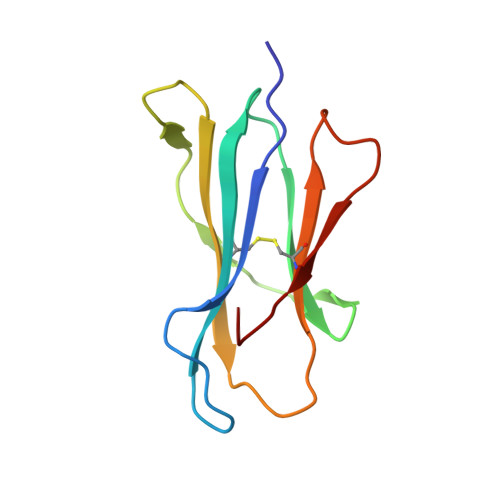
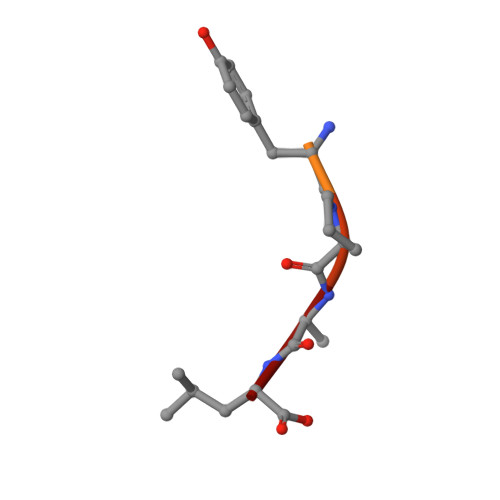
The crystal structure of H-2D(b) complexed with a partial peptide epitope suggests a major histocompatibility complex class I assembly intermediate.
Glithero, A., Tormo, J., Doering, K., Kojima, M., Jones, E.Y., Elliott, T.(2006) J Biol Chem 281: 12699-12704
- PubMed: 16478731
- DOI: https://doi.org/10.1074/jbc.M511683200
- Primary Citation of Related Structures:
2CII - PubMed Abstract:
In the absence of bound peptide ligands, major histocompatibility complex (MHC) class I molecules are unstable. In an attempt to determine the minimum requirement for peptide-dependent MHC class I stabilization, we have used short synthetic peptides derived from the Sendai virus nucleoprotein epitope (residues 324-332, 1FAPGNYPAL9) to promote its folding in vitro of H-2D(b). We found that H-2D(b) can be stabilized by the pentapeptide 5NYPAL9, which is equivalent to the C-terminal portion of the optimal nonapeptide and includes both the P5 and P9 anchor residues. We have crystallized the complex of the H-2D(b) molecule with the pentamer and determined the structure to show how a quasi-stable MHC class I molecule can be formed by occupancy of a single binding pocket in the peptide-binding groove.
Organizational Affiliation:
Nuffield Department of Clinical Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 4RU, United Kingdom.