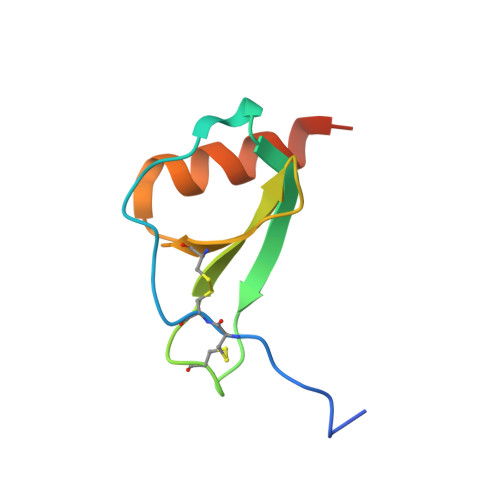
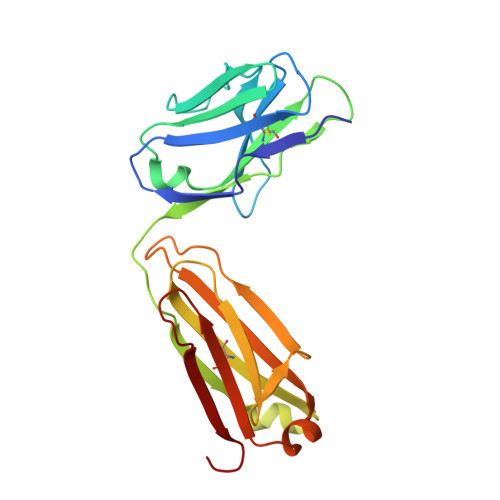
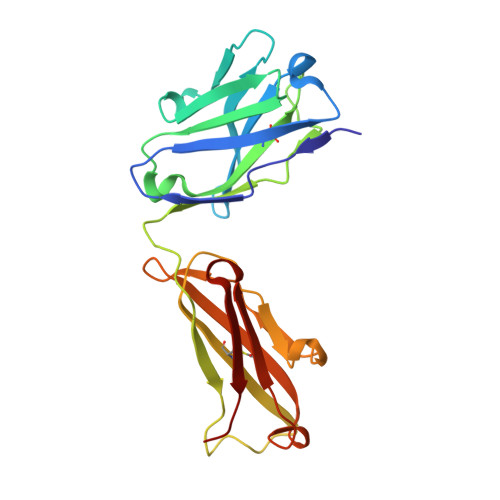
Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody.
Reid, C., Rushe, M., Jarpe, M., van Vlijmen, H., Dolinski, B., Qian, F., Cachero, T.G., Cuervo, H., Yanachkova, M., Nwankwo, C., Wang, X., Etienne, N., Garber, E., Bailly, V., de Fougerolles, A., Boriack-Sjodin, P.A.(2006) Protein Eng Des Sel 19: 317-324
- PubMed: 16682434
- DOI: https://doi.org/10.1093/protein/gzl015
- Primary Citation of Related Structures:
2BDN - PubMed Abstract:
Monocyte chemoattractant proteins (MCPs) are cytokines that direct immune cells bearing appropriate receptors to sites of inflammation or injury and are therefore attractive therapeutic targets for inhibitory molecules. 11K2 is a blocking mouse monoclonal antibody active against several human and murine MCPs. A 2.5 A structure of the Fab fragment of this antibody in complex with human MCP-1 has been solved. The Fab blocks CCR2 receptor binding to MCP-1 through an adjacent but distinct binding site. The orientation of the Fab indicates that a single MCP-1 dimer will bind two 11K2 antibodies. Several key residues on the antibody and on human MCPs were predicted to be involved in antibody selectivity. Mutational analysis of these residues confirms their involvement in the antibody-chemokine interaction. In addition to mutations that decreased or disrupted binding, one antibody mutation resulted in a 70-fold increase in affinity for human MCP-2. A key residue missing in human MCP-3, a chemokine not recognized by the antibody, was identified and engineering the preferred residue into the chemokine conferred binding to the antibody.
Organizational Affiliation:
Department of Research, Biogen Idec, Inc. 12 Cambridge Center, Cambridge, MA 02142, USA.