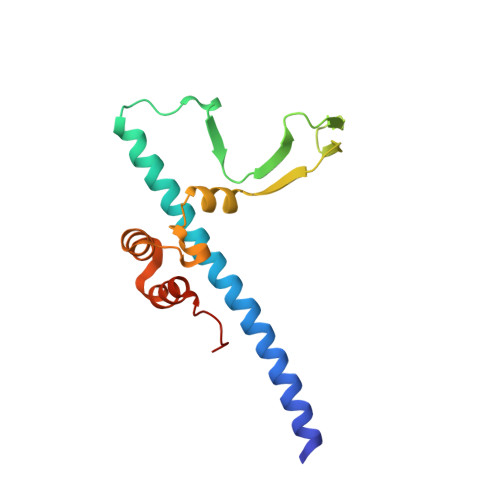
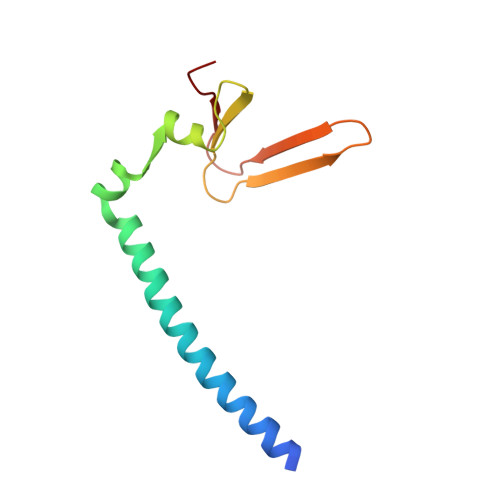
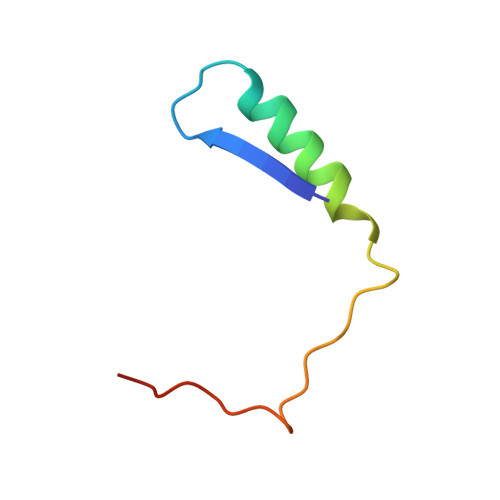
Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release.
Rubin, S.M., Gall, A.L., Zheng, N., Pavletich, N.P.(2005) Cell 123: 1093-1106
- PubMed: 16360038
- DOI: https://doi.org/10.1016/j.cell.2005.09.044
- Primary Citation of Related Structures:
2AZE - PubMed Abstract:
The retinoblastoma (Rb) protein negatively regulates the G1-S transition by binding to the E2F transcription factors, until cyclin-dependent kinases phosphorylate Rb, causing E2F release. The Rb pocket domain is necessary for E2F binding, but the Rb C-terminal domain (RbC) is also required for growth suppression. Here we demonstrate a high-affinity interaction between RbC and E2F-DP heterodimers shared by all Rb and E2F family members. The crystal structure of an RbC-E2F1-DP1 complex reveals an intertwined heterodimer in which the marked box domains of both E2F1 and DP1 contact RbC. We also demonstrate that phosphorylation of RbC at serines 788 and 795 destabilizes one set of RbC-E2F-DP interactions directly, while phosphorylation at threonines 821 and 826 induces an intramolecular interaction between RbC and the Rb pocket that destabilizes the remaining interactions indirectly. Our findings explain the requirement of RbC for high-affinity E2F binding and growth suppression and establish a mechanism for the regulation of Rb-E2F association by phosphorylation.
Organizational Affiliation:
Structural Biology Program and Howard Hughes Medical Institute, Memorial Sloan-Kettering Cancer Center, NY 10021, USA.