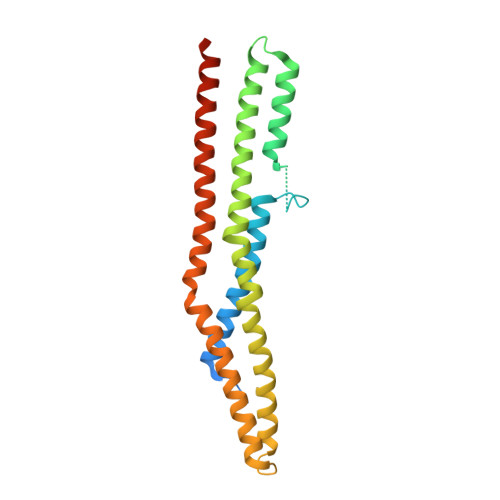
Crystal Structure of the Endophilin-A1 BAR Domain.
Weissenhorn, W.(2005) J Mol Biol 351: 653-661
- PubMed: 16023669
- DOI: https://doi.org/10.1016/j.jmb.2005.06.013
- Primary Citation of Related Structures:
1ZWW - PubMed Abstract:
Endophilin has been implicated in the retrieval of membrane via endocytosis of clathrin-coated vesicles, which is crucial for the maintenance of neurotransmitter exocytosis during stimulation; both exocytosis and endocytosis are regulated by intracellular calcium levels. Here, we present the 2.3 A crystal structure of the endophilin-A1 BAR domain, which has been suggested to function in inducing and sensing membrane curvature at the site of endocytosis. Endo-BAR folds into a crescent-shaped dimer composed of two elongated, three-helix bundles. Two additional domains of 30 residues each, inserted into helix 1 at the center of the concave side of the dimer, may interfere with the proposed mode of BAR domain membrane interaction. In addition, the dimer binds 11 divalent cadmium ions in the crystal mostly with typical Ca2+ co-ordination spheres. The endophilin-1A BAR domain thus constitutes a new variant of a BAR domain, and it may link endophilin-1A BAR function to calcium regulation of endocytosis.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), 6 rue Jules Horowitz, 38042 Grenoble, France. weissen@embl-grenoble.fr