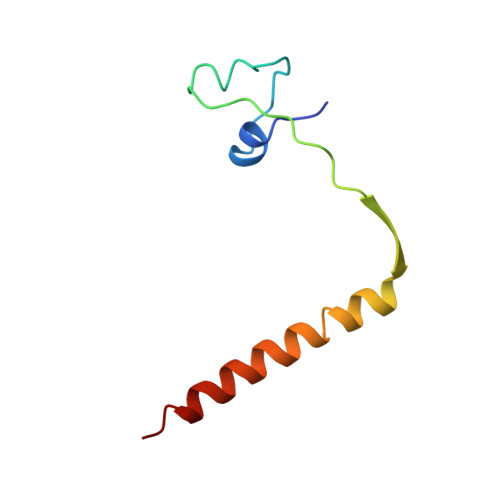
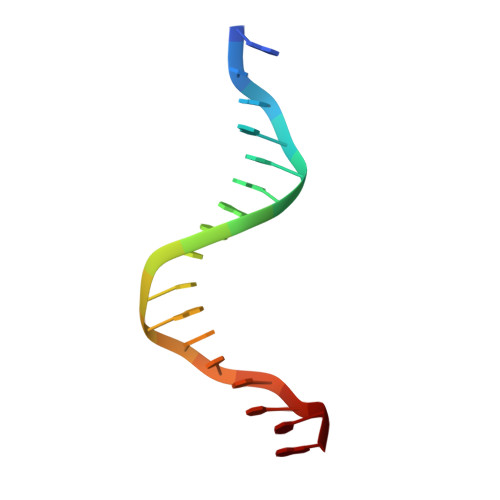
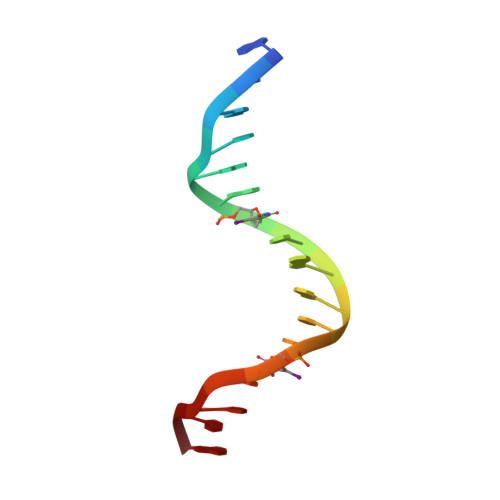
Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster.
Swaminathan, K., Flynn, P., Reece, R.J., Marmorstein, R.(1997) Nat Struct Biol 4: 751-759
- PubMed: 9303004
- DOI: https://doi.org/10.1038/nsb0997-751
- Primary Citation of Related Structures:
1ZME - PubMed Abstract:
PUT3 is a member of a family of at least 79 fungal transcription factors that contain a six-cysteine, two-zinc domain called a 'Zn2Cys6 binuclear cluster'. We have determined the crystal structure of the DNA binding region from the PUT3 protein bound to its cognate DNA target. The structure reveals that the PUT3 homodimer is bound asymmetrically to the DNA site. This asymmetry orients a beta-strand from one protein subunit into the minor groove of the DNA resulting in a partial amino acid-base pair intercalation and extensive direct and water-mediated protein interactions with the minor groove of the DNA. These interactions facilitate a sequence dependent kink at the centre of the DNA site and specify the intervening base pairs separating two DNA half-sites that are contacted in the DNA major groove. A comparison with the GAL4-DNA and PPR1-DNA complexes shows how a family of related DNA binding proteins can use a diverse set of mechanisms to discriminate between the base pairs separating conserved DNA half-sites.
Organizational Affiliation:
Wistar Institute, University of Pennsylvania, Philadelphia 19104, USA.