Structural Insights into the Mechanism of Nuclease A, a beta beta alpha Metal Nuclease from Anabaena.
Ghosh, M., Meiss, G., Pingoud, A., London, R.E., Pedersen, L.C.(2005) J Biol Chem 280: 27990-27997
- PubMed: 15897201
- DOI: https://doi.org/10.1074/jbc.M501798200
- Primary Citation of Related Structures:
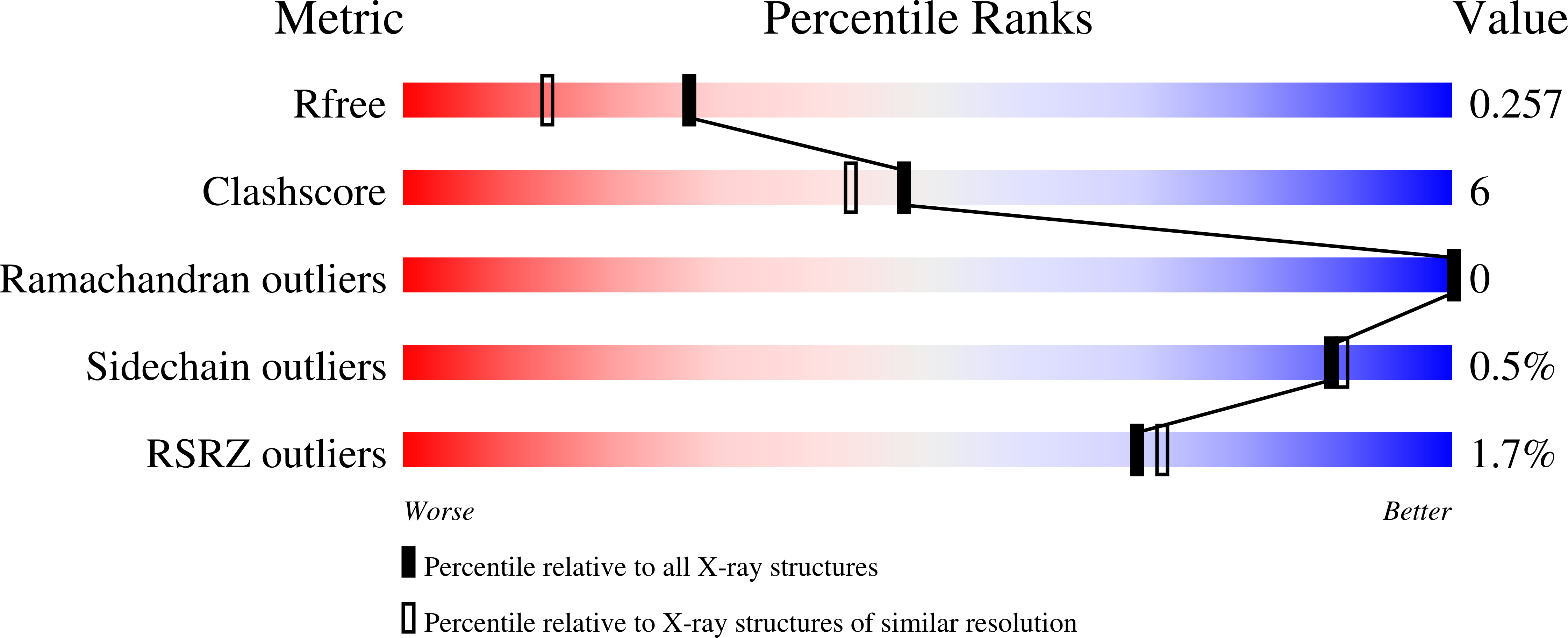
1ZM8 - PubMed Abstract:
Nuclease A (NucA) is a nonspecific endonuclease from Anabaena sp. capable of degrading single- and double-stranded DNA and RNA in the presence of divalent metal ions. We have determined the structure of the delta(2-24),D121A mutant of NucA in the presence of Zn2+ and Mn2+ (PDB code 1ZM8). The mutations were introduced to remove the N-terminal signal peptide and to reduce the activity of the nonspecific nuclease, thereby reducing its toxicity to the Escherichia coli expression system. NucA contains a betabeta alpha metal finger motif and a hydrated Mn2+ ion at the active site. Unexpectedly, NucA was found to contain additional metal binding sites approximately 26 A apart from the catalytic metal binding site. A structural comparison between NucA and the closest analog for which structural data exist, the Serratia nuclease, indicates several interesting differences. First, NucA is a monomer rather than a dimer. Second, there is an unexpected structural homology between the N-terminal segments despite a poorly conserved sequence, which in Serratia includes a cysteine bridge thought to play a regulatory role. In addition, although a sequence alignment had suggested that NucA lacks a proposed catalytic residue corresponding to Arg57 in Serratia, the structure determined here indicates that Arg93 in NucA is positioned to fulfill this role. Based on comparison with DNA-bound nuclease structures of the betabeta alpha metal finger nuclease family and available mutational data on NucA, we propose that His124 acts as a catalytic base, and Arg93 participates in the catalysis possibly through stabilization of the transition state.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina 27709, USA.