The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
Arolas, J.L., Popowicz, G.M., Lorenzo, J., Sommerhoff, C.P., Huber, R., Aviles, F.X., Holak, T.A.(2005) J Mol Biol 350: 489-498
- PubMed: 15961103
- DOI: https://doi.org/10.1016/j.jmb.2005.05.015
- Primary Citation of Related Structures:
1ZLH, 1ZLI - PubMed Abstract:
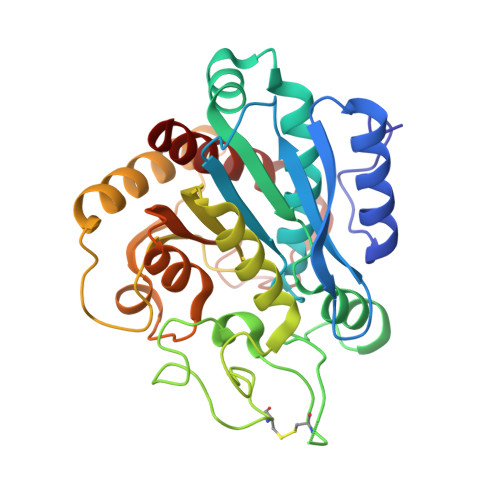
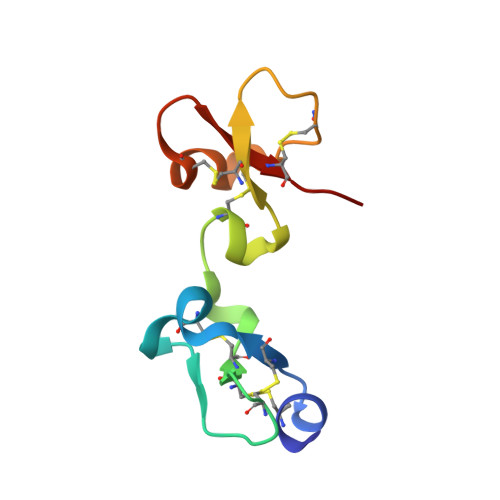
The tick carboxypeptidase inhibitor (TCI) is a proteinaceous inhibitor of metallo-carboxypeptidases present in the blood-sucking tick Rhipicephalus bursa. The three-dimensional crystal structures of recombinant TCI bound to bovine carboxypeptidase A and to human carboxypeptidase B have been determined and refined at 1.7 A and at 2.0 A resolution, respectively. TCI consists of two domains that are structurally similar despite the low degree of sequence homology. The domains, each consisting of a short alpha-helix followed by a small twisted antiparallel beta-sheet, show a high level of structural homology to proteins of the beta-defensin-fold family. TCI anchors to the surface of mammalian carboxypeptidases in a double-headed manner not previously seen for carboxypeptidase inhibitors: the last three carboxy-terminal amino acid residues interact with the active site of the enzyme in a way that mimics substrate binding, and the N-terminal domain binds to an exosite distinct from the active-site groove. The structures of these complexes should prove valuable in the applications of TCI as a thrombolytic drug and as a basis for the design of novel bivalent carboxypeptidase inhibitors.
Organizational Affiliation:
Institut de Biotecnologia i Biomedicina and Departament de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain.