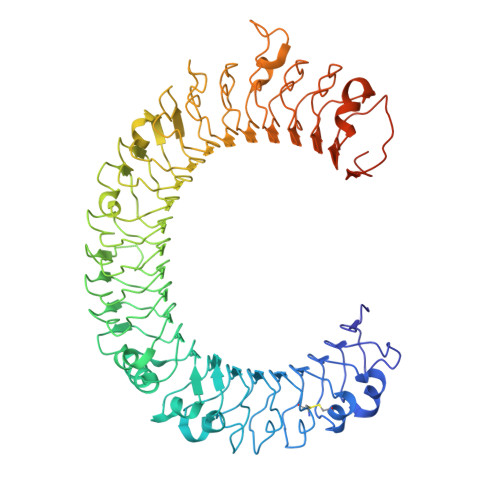
Crystal structure of human toll-like receptor 3 (TLR3) ectodomain.
Choe, J., Kelker, M.S., Wilson, I.A.(2005) Science 309: 581-585
- PubMed: 15961631
- DOI: https://doi.org/10.1126/science.1115253
- Primary Citation of Related Structures:
1ZIW - PubMed Abstract:
Toll-like receptors (TLRs) play key roles in activating immune responses during infection. The human TLR3 ectodomain structure at 2.1 angstroms reveals a large horseshoe-shaped solenoid assembled from 23 leucine-rich repeats (LRRs). Asparagines conserved in the 24-residue LRR motif contribute extensive hydrogen-bonding networks for solenoid stabilization. TLR3 is largely masked by carbohydrate, but one face is glycosylation-free, which suggests its potential role in ligand binding and oligomerization. Highly conserved surface residues and a TLR3-specific LRR insertion form a homodimer interface in the crystal, whereas two patches of positively charged residues and a second insertion would provide an appropriate binding site for double-stranded RNA.
Organizational Affiliation:
Department of Molecular Biology and Skaggs Institute for Chemical Biology, Scripps Research Institute (TSRI), 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.