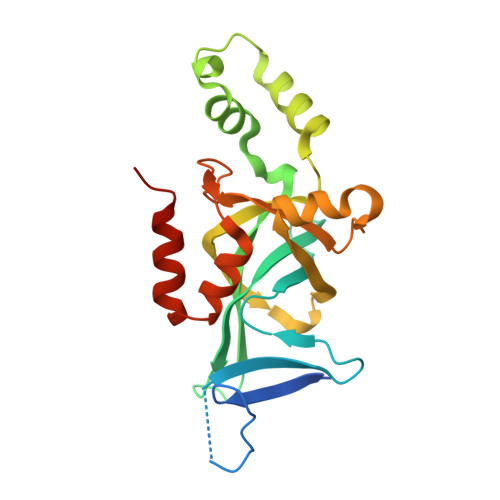
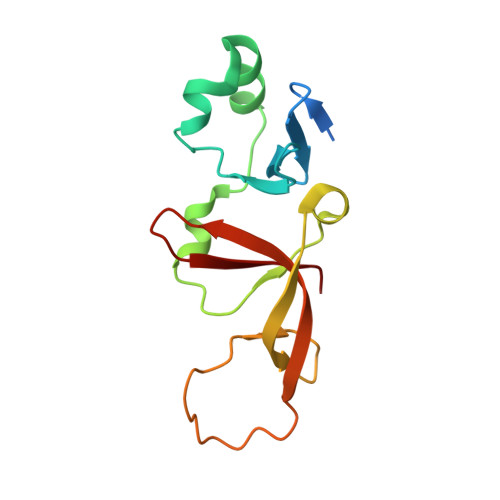
Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing.
Hou, Z., Bernstein, D.A., Fox, C.A., Keck, J.L.(2005) Proc Natl Acad Sci U S A 102: 8489-8494
- PubMed: 15932939
- DOI: https://doi.org/10.1073/pnas.0503525102
- Primary Citation of Related Structures:
1Z1A, 1ZHI - PubMed Abstract:
The Sir1 protein plays a key role in establishing a silent chromatin structure at the cryptic mating-type loci HMR and HML in Saccharomyces cerevisiae by interacting with the bromo-adjacent homology (BAH) domain of the Orc1p subunit of the origin recognition complex (ORC). Here, we present the high-resolution crystal structures of the ORC interaction region (OIR) of Sir1p and that of the complex formed between the OIR and BAH domains. Amino acids within the OIR previously shown to be required for a Sir1p/ORC interaction are presented on a conserved, convex surface that forms a complementary interface with a concave region of the Orc1 BAH domain that is critical for transcriptional silencing. The OIR/BAH interaction surface comprises a network of hydrophobic and polar/ionic interactions between discrete structural modules in each protein and involves several residues that were not implicated in previous studies. These data provide important structural insights into a protein-protein interaction critical for the formation of a specialized chromatin domain within eukaryotic chromosomes.
Organizational Affiliation:
Department of Biomolecular Chemistry, University of Wisconsin Medical School, Medical Sciences Center, 1300 University Avenue, Madison, WI 53706-1532, USA.