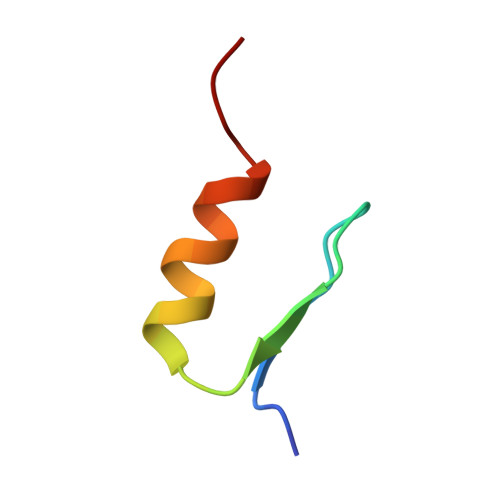
Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet.
Neuhaus, D., Nakaseko, Y., Schwabe, J.W., Klug, A.(1992) J Mol Biol 228: 637-651
- PubMed: 1453468
- DOI: https://doi.org/10.1016/0022-2836(92)90846-c
- Primary Citation of Related Structures:
1ZFD - PubMed Abstract:
This paper describes the detailed three-dimensional structures of two zinc-finger domains from the yeast transcription factor SWI5, calculated using the results of the n.m.r. experiments described in the accompanying paper. The structure of finger 2 is essentially similar to those previously obtained by others for isolated, synthetic single zinc-finger domains in solution, and for the three zinc-finger peptide Zif268 in its crystalline complex with DNA. The N-terminal half of the sequence forms a two-stranded, irregular beta-sheet containing both of the metal-binding cysteine residues, while the remainder of the structure forms a helix. Approximately the first half of this helix is alpha-helical, whereas the C-terminal portion, including the two metal-binding histidine residues, is 3(10) helical. Four invariant hydrophobic residues form a core to the structure. In contrast to all previously described structures of zinc-finger domains, finger 1 has an additional strand in the beta-sheet, formed by residues N-terminal to the formal start of the finger motif. This additional strand plays a role in stabilising the folded form of finger 1, since a two-finger peptide lacking the N-terminal residues showed folded structure in finger 2 but not in finger 1.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, England.