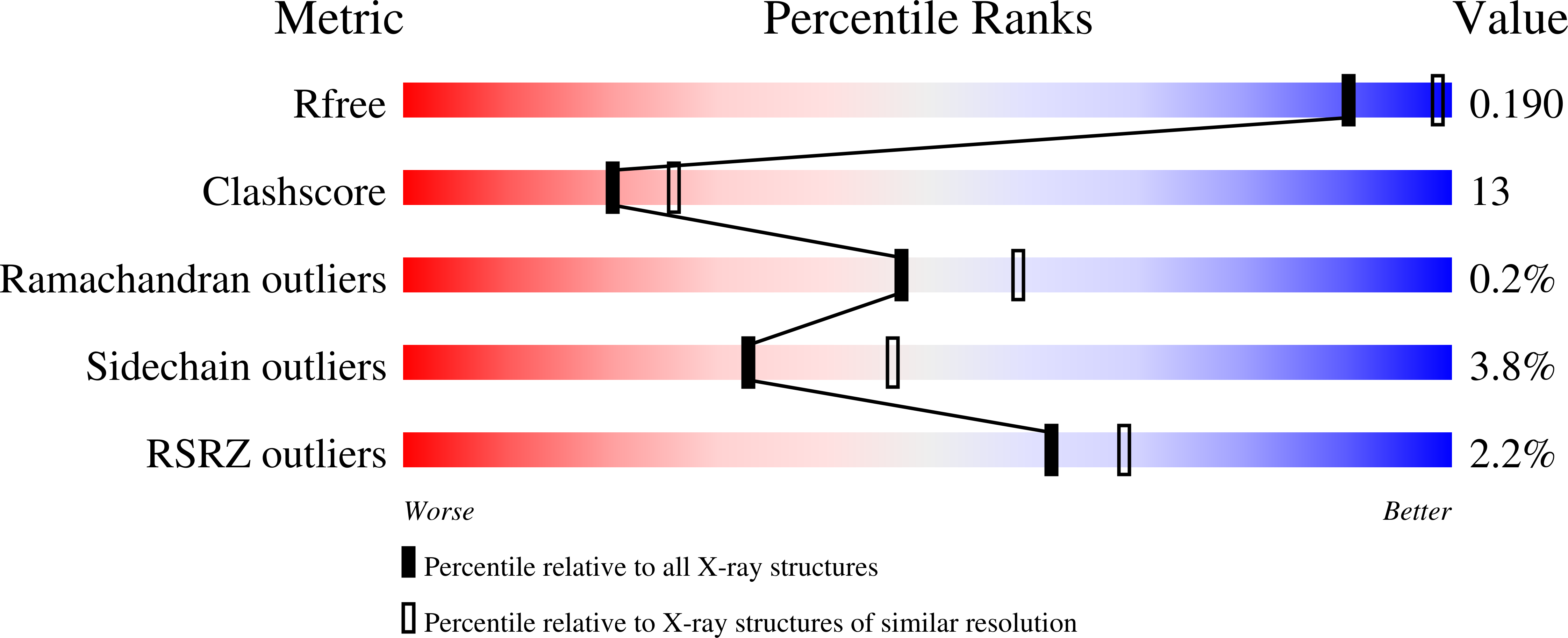
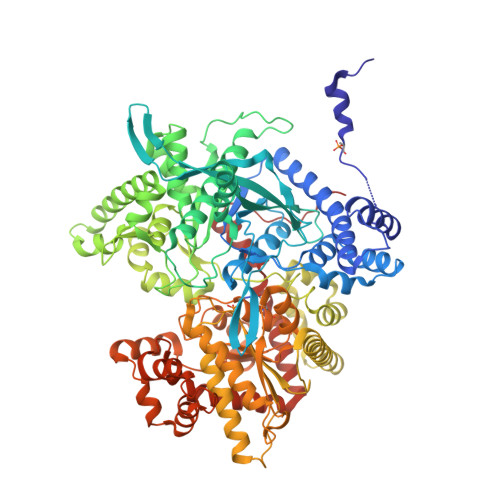
The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Lukacs, C.M., Oikonomakos, N.G., Crowther, R.L., Hong, L.N., Kammlott, R.U., Levin, W., Li, S., Liu, C.M., Lucas-McGady, D., Pietranico, S., Reik, L.(2006) Proteins 63: 1123-1126
- PubMed: 16523484
- DOI: https://doi.org/10.1002/prot.20939
- Primary Citation of Related Structures:
1Z8D
Organizational Affiliation:
Roche Pharmaceuticals, Discovery Chemistry, F. Hoffmann-La Roche, Nutley, New Jersey 07110, USA. christine.lukacs@roche.com