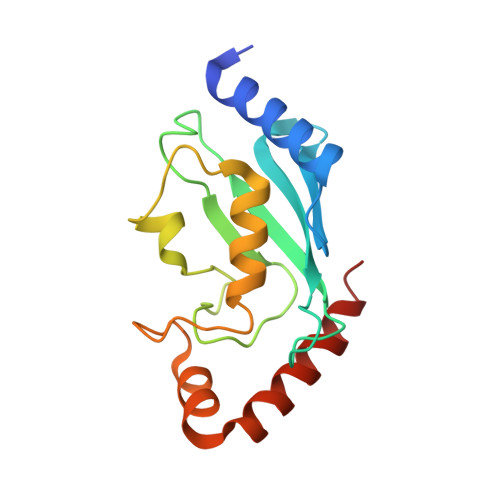
Protein crystal growth improvement leading to the 2.5A crystallographic structure of ubiquitin-conjugating enzyme (ubc-1) from Caenorhabditis elegans
Gavira, J.A., DiGiammarino, E., Tempel, W., Toh, D., Liu, Z.J., Wang, B.C., Meehan, E., Ng, J.D.To be published.