Structural Analysis of Dispersin B, a Biofilm-releasing Glycoside Hydrolase from the Periodontopathogen Actinobacillus actinomycetemcomitans.
Ramasubbu, N., Thomas, L.M., Ragunath, C., Kaplan, J.B.(2005) J Mol Biol 349: 475-486
- PubMed: 15878175
- DOI: https://doi.org/10.1016/j.jmb.2005.03.082
- Primary Citation of Related Structures:
1YHT - PubMed Abstract:
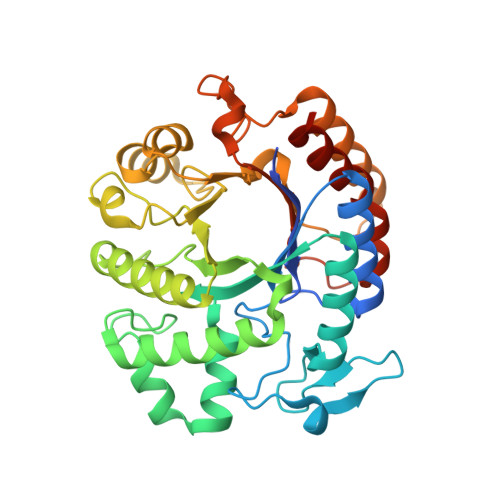
Bacteria in a biofilm are enmeshed in a self-synthesized extracellular polysaccharide matrix that holds the bacteria together in a mass and firmly attaches the bacterial mass to the underlying surface. A major component of the extracellular polysaccharide matrix in several phylogenetically diverse bacteria is PGA, a linear polymer of N-acetylglucosamine residues in beta(1,6)-linkage. PGA is produced by the Gram-negative periodontopathogen Actinobacillus actinomycetemcomitans as well as by the Gram-positive device-associated pathogen Staphylococcus epidermidis. We recently reported that A.actinomycetemcomitans produces a soluble glycoside hydrolase named dispersin B, which degrades PGA. Here, we present the crystal structure of dispersin B at 2.0A in complex with a glycerol and an acetate ion at the active site. The enzyme crystallizes in the orthorhombic space group C222(1) with cell dimensions a=41.02A, b=86.13A, c=185.77A. The core of the enzyme consists a (beta/alpha)(8) barrel topology similar to other beta-hexosaminidases but significant differences exist in the arrangement of loops hovering in the vicinity of the active site. The location and interactions of the glycerol and acetate moieties in conjunction with the sequence analysis suggest that dispersin B cleaves beta(1,6)-linked N-acetylglucosamine polymer using a catalytic machinery similar to other family 20 hexosaminidases which cleave beta(1,4)-linked N-acetylglucosamine residues.
Organizational Affiliation:
Department of Oral Biology, University of Medicine and Dentistry of New Jersey, Newark, NJ 07103, USA. ramasun1@umdnj.edu