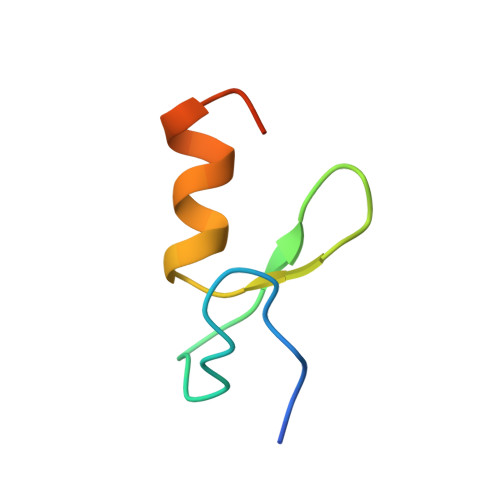
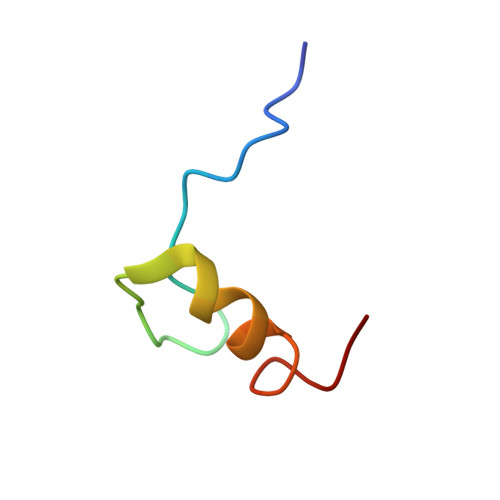
Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Liew, C.K., Simpson, R.J.Y., Kwan, A.H.Y., Crofts, L.A., Loughlin, F.E., Matthews, J.M., Crossley, M., Mackay, J.P.(2005) Proc Natl Acad Sci U S A 102: 583-588
- PubMed: 15644435
- DOI: https://doi.org/10.1073/pnas.0407511102
- Primary Citation of Related Structures:
1Y0J - PubMed Abstract:
GATA-1 and friend of GATA (FOG) are zinc-finger transcription factors that physically interact to play essential roles in erythroid and megakaryocytic development. Several naturally occurring mutations in the GATA-1 gene that alter the FOG-binding domain have been reported. The mutations are associated with familial anemias and thrombocytopenias of differing severity. To elucidate the molecular basis for the GATA-1/FOG interaction, we have determined the three-dimensional structure of a complex comprising the interaction domains of these proteins. The structure reveals how zinc fingers can act as protein recognition motifs. Details of the architecture of the contact domains and their physical properties provide a molecular explanation for how the GATA-1 mutations contribute to distinct but related genetic diseases.
Organizational Affiliation:
School of Molecular and Microbial Biosciences, University of Sydney, Sydney, New South Wales 2006, Australia.