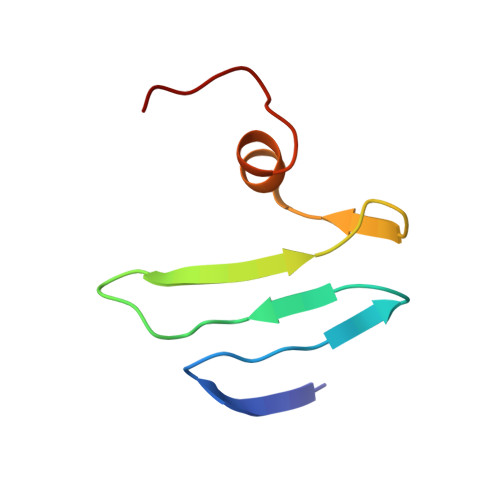
Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
Gutierrez, P., Li, Y., Osborne, M.J., Pomerantseva, E., Liu, Q., Gehring, K.(2005) J Bacteriol 187: 3496-3501
- PubMed: 15866937
- DOI: https://doi.org/10.1128/JB.187.10.3496-3501.2005
- Primary Citation of Related Structures:
1Y00 - PubMed Abstract:
The carbon storage regulator A (CsrA) is a protein responsible for the repression of a variety of stationary-phase genes in bacteria. In this work, we describe the nuclear magnetic resonance (NMR)-based structure of the CsrA dimer and its RNA-binding properties. CsrA is a dimer of two identical subunits, each composed of five strands, a small alpha-helix and a flexible C terminus. NMR titration experiments suggest that the beta1-beta2 and beta3-beta4 loops and the C-terminal helix are important elements in RNA binding. Even though the beta3-beta4 loop contains a highly conserved RNA-binding motif, GxxG, typical of KH domains, our structure excludes CsrA from being a member of this protein family, as previously suggested. A mechanism for the recognition of mRNAs downregulated by CsrA is proposed.
Organizational Affiliation:
Department of Biochemistry, McGill University, 3655 Promenade Sir William Osler, Montreal, Quebec, Canada H3G 1Y6.