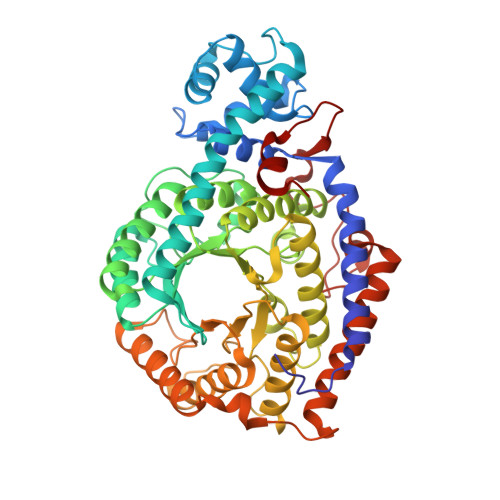
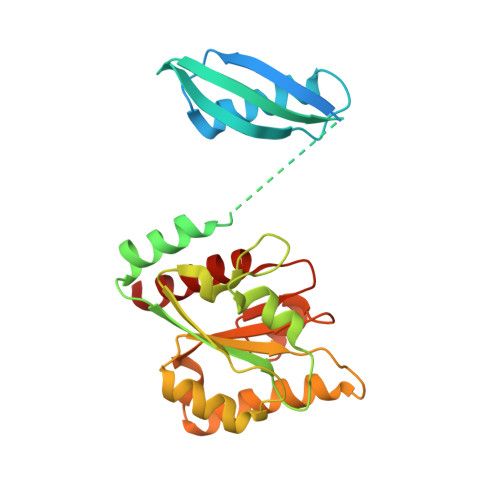
A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase.
Berkovitch, F., Behshad, E., Tang, K.H., Enns, E.A., Frey, P.A., Drennan, C.L.(2004) Proc Natl Acad Sci U S A 101: 15870-15875
- PubMed: 15514022
- DOI: https://doi.org/10.1073/pnas.0407074101
- Primary Citation of Related Structures:
1XRS - PubMed Abstract:
Lysine 5,6-aminomutase is an adenosylcobalamin and pyridoxal-5'-phosphate-dependent enzyme that catalyzes a 1,2 rearrangement of the terminal amino group of dl-lysine and of l-beta-lysine. We have solved the x-ray structure of a substrate-free form of lysine-5,6-aminomutase from Clostridium sticklandii. In this structure, a Rossmann domain covalently binds pyridoxal-5'-phosphate by means of lysine 144 and positions it into the putative active site of a neighboring triosephosphate isomerase barrel domain, while simultaneously positioning the other cofactor, adenosylcobalamin, approximately 25 A from the active site. In this mode of pyridoxal-5'-phosphate binding, the cofactor acts as an anchor, tethering the separate polypeptide chain of the Rossmann domain to the triosephosphate isomerase barrel domain. Upon substrate binding and transaldimination of the lysine-144 linkage, the Rossmann domain would be free to rotate and bring adenosylcobalamin, pyridoxal-5'-phosphate, and substrate into proximity. Thus, the structure embodies a locking mechanism to keep the adenosylcobalamin out of the active site and prevent radical generation in the absence of substrate.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.