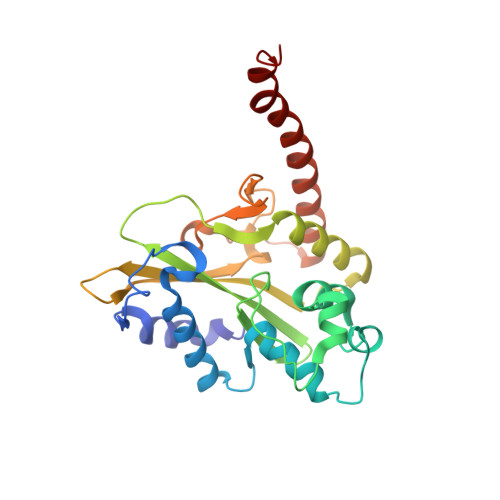
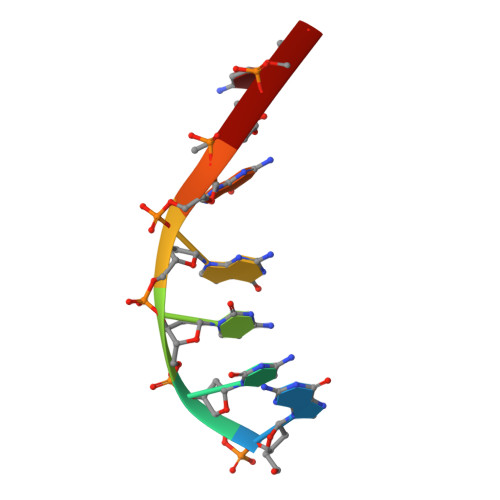
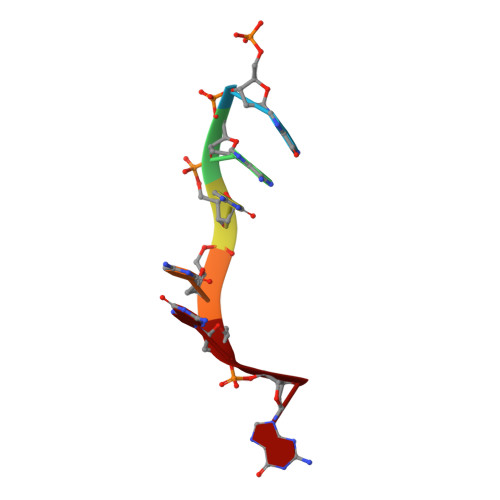
Mechanistic Insights from the Structures of HincII Bound to Cognate DNA Cleaved from Addition of Mg(2+) and Mn(2+)
Etzkorn, C., Horton, N.C.(2004) J Mol Biol 343: 833-849
- PubMed: 15476804
- DOI: https://doi.org/10.1016/j.jmb.2004.08.082
- Primary Citation of Related Structures:
1XHU, 1XHV - PubMed Abstract:
The three-dimensional X-ray crystal structures of HincII bound to cognate DNA containing GTCGAC and Mn(2+) or Mg(2+), at 2.50A and 2.95A resolution, respectively, are presented. In both structures, the DNA is found cleaved, and the positions of the active-site groups, cleaved phosphate group, and 3' oxygen atom of the leaving group are in very similar positions. Two highly occupied Mn(2+) positions are found in each active site of the four crystallographically independent subunit copies in the HincII/DNA/Mn(2+) structure. The manganese ion closest to the previously identified single Ca(2+) position of HincII is shifted 1.7A and has lost direct ligation to the active-site aspartate residue, Asp127. A Mn(2+)-ligated water molecule in a position analogous to that seen in the HincII/DNA/Ca(2+) structure, and proposed to be the attacking nucleophile, is beyond hydrogen bonding distance from the active-site lysine residue, Lys129, but remains within hydrogen bonding distance from the proRp oxygen atom of the phosphate group 3' to the scissile phosphate group. In addition, the position of the cleaved phosphate group is on the opposite side of the axis connecting the two metal ions relative to that found in the BamHI/product DNA/Mn(2+) structure. Mechanistic implications are discussed, and a model for the two-metal-ion mechanism of DNA cleavage by HincII is proposed.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, University of Arizona, Tucson, AZ 85721, USA.