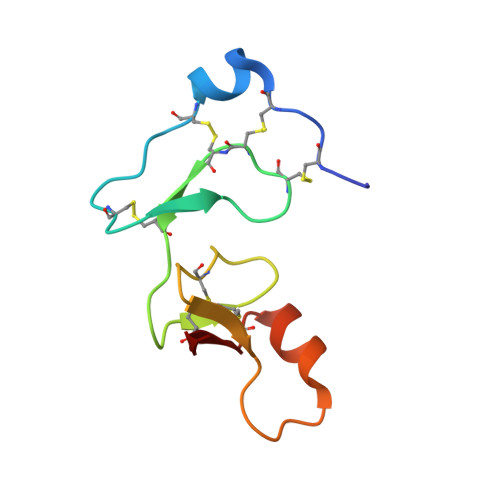
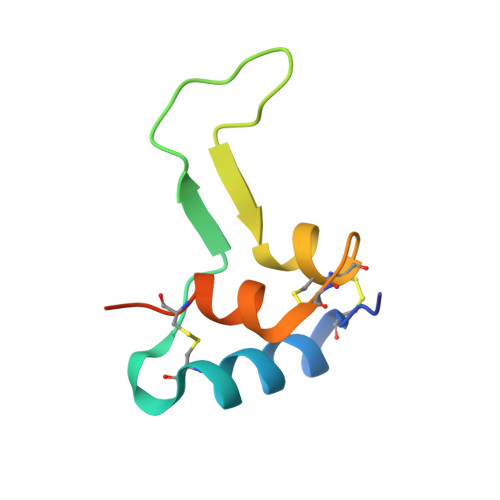
Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Siwanowicz, I., Popowicz, G.M., Wisniewska, M., Huber, R., Kuenkele, K.P., Lang, K., Engh, R.A., Holak, T.A.(2005) Structure 13: 155-167
- PubMed: 15642270
- DOI: https://doi.org/10.1016/j.str.2004.11.009
- Primary Citation of Related Structures:
1WQJ - PubMed Abstract:
Insulin-like growth factor binding proteins (IGFBPs) control the extracellular distribution, function, and activity of IGFs. Here, we report an X-ray structure of the binary complex of IGF-I and the N-terminal domain of IGFBP-4 (NBP-4, residues 3-82) and a model of the ternary complex of IGF-I, NBP-4, and the C-terminal domain (CBP-4, residues 151-232) derived from diffraction data with weak definition of the C-terminal domain. These structures show how the IGFBPs regulate IGF signaling. Key features of the structures include (1) a disulphide bond ladder that binds to IGF and partially masks the IGF residues responsible for type 1 IGF receptor (IGF-IR) binding, (2) the high-affinity IGF-I interaction site formed by residues 39-82 in a globular fold, and (3) CBP-4 interactions. Although CBP-4 does not bind individually to either IGF-I or NBP-4, in the ternary complex, CBP-4 contacts both and also blocks the IGF-IR binding region of IGF-I.
Organizational Affiliation:
Max Planck Institut für Biochemie, D-82152 Martinsried, Germany.