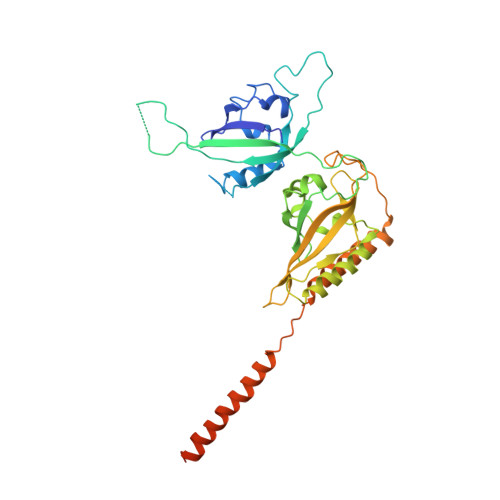
Crystal Structure and Interactions of the Pas Repeat Region of the Drosophila Clock Protein Period
Yildiz, O., Doi, M., Yujnovsky, I., Cardone, L., Berndt, A., Hennig, S., Schulze, S., Urbanke, C., Sassone-Corsi, P., Wolf, E.(2005) Mol Cell 17: 69
- PubMed: 15629718
- DOI: https://doi.org/10.1016/j.molcel.2004.11.022
- Primary Citation of Related Structures:
1WA9 - PubMed Abstract:
PERIOD proteins are central components of the Drosophila and mammalian circadian clock. Their function is controlled by daily changes in synthesis, cellular localization, phosphorylation, degradation, as well as specific interactions with other clock components. Here we present the crystal structure of a Drosophila PERIOD (dPER) fragment comprising two tandemly organized PAS (PER-ARNT-SIM) domains (PAS-A and PAS-B) and two additional C-terminal alpha helices (alphaE and alphaF). Our analysis reveals a noncrystallographic dPER dimer mediated by intermolecular interactions of PAS-A with PAS-B and helix alphaF. We show that alphaF is essential for dPER homodimerization and that the PAS-A-alphaF interaction plays a crucial role in dPER clock function, as it is affected by the 29 hr long-period perL mutation.
Organizational Affiliation:
Department of Structural Biology, Max-Planck-Institute for Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.