Crystal Structure of Yeast Ypr118W, a Methylthioribose-1-Phosphate Isomerase Related to Regulatory Eif2B Subunits
Bumann, M., Djafarzadeh, S., Oberholzer, A.E., Bigler, P., Altmann, M., Trachsel, H., Baumann, U.(2004) J Biol Chem 279: 37087
- PubMed: 15215245
- DOI: https://doi.org/10.1074/jbc.M404458200
- Primary Citation of Related Structures:
1W2W - PubMed Abstract:
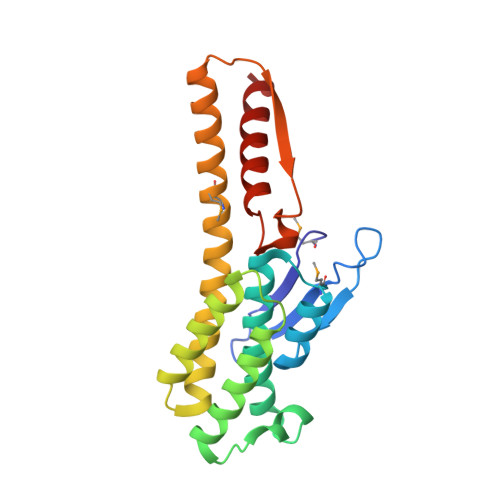
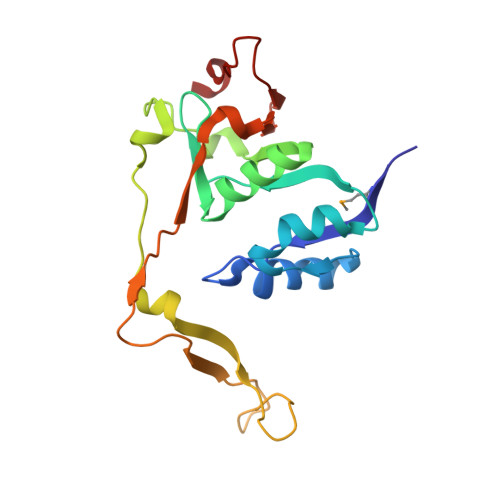
Ypr118w is a non-essential, low copy number gene product from Saccharomyces cerevisiae. It belongs to the PFAM family PF01008, which contains the alpha-, beta-, and delta-subunits of eukaryotic translation initiation factor eIF2B, as well as proteins of unknown function from all three kingdoms. Recently, one of those latter proteins from Bacillus subtilis has been characterized as a 5-methylthioribose-1-phosphate isomerase, an enzyme of the methionine salvage pathway. We report here the crystal structure of Ypr118w, which reveals a dimeric protein with two domains and a putative active site cleft. The C-terminal domain resembles ribose-5-phosphate isomerase from Escherichia coli with a similar location of the active site. In vivo, Ypr118w protein is required for yeast cells to grow on methylthioadenosine in the absence of methionine, showing that Ypr118w is involved in the methionine salvage pathway. The crystal structure of Ypr118w reveals for the first time the fold of a PF01008 member and allows a deeper discussion of an enzyme of the methionine salvage pathway, which has in the past attracted interest due to tumor suppression and as a target of aniprotozoal drugs.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Berne, Freiestrasse 3, Berne CH-3012.