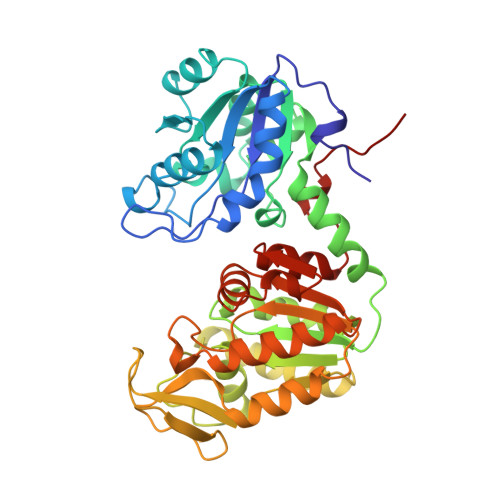
Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Auerbach, G., Huber, R., Grattinger, M., Zaiss, K., Schurig, H., Jaenicke, R., Jacob, U.(1997) Structure 5: 1475-1483
- PubMed: 9384563
- DOI: https://doi.org/10.1016/s0969-2126(97)00297-9
- Primary Citation of Related Structures:
1VPE - PubMed Abstract:
Phosphoglycerate kinase (PGK) is essential in most living cells both for ATP generation in the glycolytic pathway of aerobes and for fermentation in anaerobes. In addition, in many plants the enzyme is involved in carbon fixation. Like other kinases, PGK folds into two distinct domains, which undergo a large hinge-bending motion upon catalysis. The monomeric 45 kDa enzyme catalyzes the transfer of the C1-phosphoryl group from 1, 3-bisphosphoglycerate to ADP to form 1,3-bisphosphoglycerate to ADP to form 3-phosphoglycerate and ATP. For decades, the conformation of the enzyme during catalysis has been enigmatic. The crystal structure of PGK from the hyperthermophilic organism Thermotoga maritima (TmPGK) represents the first structure of an extremely thermostable PGK. It adds to a series of four known crystal structures of PGKs from mesophilic via moderately thermophilic to a hyperthermophilic organism, allowing a detailed analysis of possible structural determinants of thermostability.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abt. Strukturforschung, 82152, Martinsried, Germany. auerbach@biochem.mpg.de