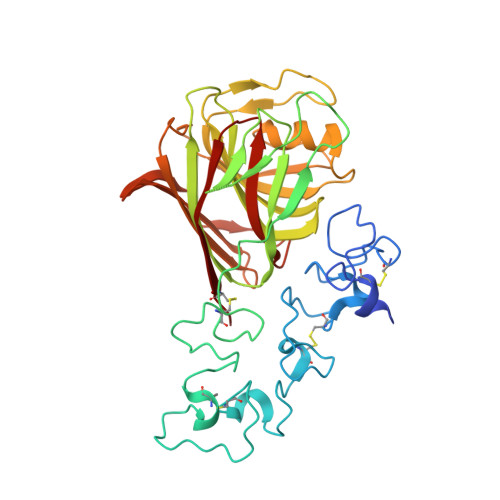
Structure of a Thrombospondin C-Terminal Fragment Reveals a Novel Calcium Core in the Type 3 Repeats
Kvansakul, M., Adams, J.C., Hohenester, E.(2004) EMBO J 23: 1223
- PubMed: 15014436
- DOI: https://doi.org/10.1038/sj.emboj.7600166
- Primary Citation of Related Structures:
1UX6 - PubMed Abstract:
Thrombospondins (TSPs) are extracellular regulators of cell-matrix interactions and cell phenotype. The most highly conserved region of all TSPs are the calcium-binding type 3 (T3) repeats and the C-terminal globular domain (CTD). The crystal structure of a cell-binding TSP-1 fragment, spanning three T3 repeats and the CTD, reveals a compact assembly. The T3 repeats lack secondary structure and are organised around a core of calcium ions; two DxDxDGxxDxxD motifs per repeat each encapsulate two calcium ions in a novel arrangement. The CTD forms a lectin-like beta-sandwich and contains four strictly conserved calcium-binding sites. Disruption of the hairpin structure of T3 repeats 6 and 7 decreases protein secretion and stability. The availability for cell attachment of an RGD motif in T3 repeat 7 is modulated by calcium loading. The central architectural role of calcium explains how it is critical for the functions of the TSP C-terminal region. Mutations in the T3 repeats of TSP-5/COMP, which cause two human skeletal disorders, are predicted to disrupt the tertiary structure of the T3-CTD assembly.
Organizational Affiliation:
Department of Biological Sciences, Imperial College London, South Kensington Campus, London, UK.