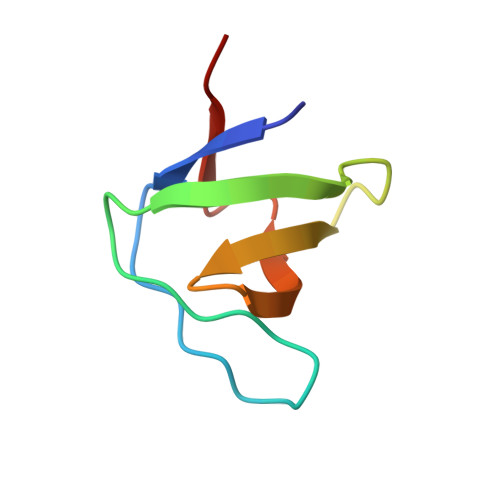
Mona/Gads Sh3C Binding to Hematopoietic Progenitor Kinase 1 (Hpk1) Combines an Atypical SH3 Binding Motif, R/Kxxk, with a Classical Pxxp Motif Embedded in a Polyproline Type II (Ppii) Helix
Lewitzky, M., Harkiolaki, M., Domart, M.C., Jones, E., Feller, S.M.(2004) J Biol Chem 279: 28724
- PubMed: 15100220
- DOI: https://doi.org/10.1074/jbc.M402745200
- Primary Citation of Related Structures:
1UTI - PubMed Abstract:
Hematopoietic progenitor kinase 1 (HPK1) is implicated in signaling downstream of the T cell receptor. Its non-catalytic, C-terminal half contains several prolinerich motifs, which have been shown to interact with different SH3 domain-containing adaptor proteins in vitro. One of these, Mona/Gads, was also shown to bind HPK1 in mouse T cells in vivo. The region of HPK1 that binds to the Mona/Gads C-terminal SH3 domain has been mapped and shows only very limited similarity to a recently identified high affinity binding motif in SLP-76, another T-cell adaptor. Using isothermal titration calorimetry and x-ray crystallography, the binding of the HPK1 motif to Mona/Gads SH3C has now been characterized in molecular detail. The results indicate that although charge interactions through an RXXK motif are essential for complex formation, a PXXP motif in HPK1 strongly complements binding. This unexpected binding mode therefore differs considerably from the previously described interaction of Mona/Gads SH3C with SLP-76. The crystal structure of the complex highlights the great versatility of SH3 domains, which allows interactions with very different proteins. This currently limits our ability to categorize SH3 binding properties by simple rules.
Organizational Affiliation:
Cancer Research UK Cell Signalling Group, Molecular Oncology Laboratory, Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom.