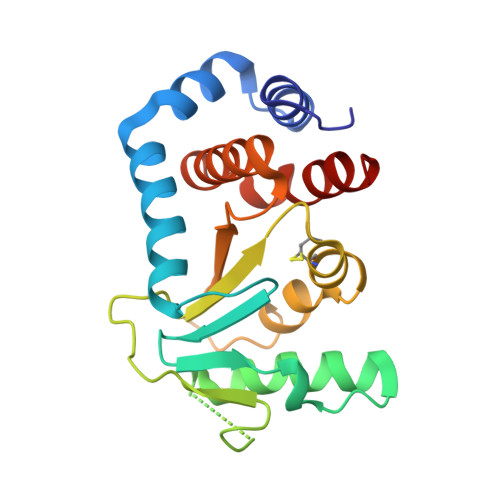
Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Manjasetty, B.A., Hennecke, J., Glockshuber, R., Heinemann, U.(2004) Acta Crystallogr D Biol Crystallogr 60: 304
- PubMed: 14747707
- DOI: https://doi.org/10.1107/S0907444903028695
- Primary Citation of Related Structures:
1UN2 - PubMed Abstract:
The thiol-disulfide oxidoreductase DsbA is required for efficient formation of disulfide bonds in the Escherichia coli periplasm. The enzyme is the strongest oxidant of the family of thioredoxin-like proteins and three-dimensional structures of both oxidized and reduced forms are known. DsbA consists of a catalytic thioredoxin-like domain and a helical domain that is inserted into the thioredoxin motif. Here, the X-ray structure of a circularly permuted variant, cpDsbA(Q100T99), is reported in which the natural termini are joined by the pentapeptide linker GGGTG, leading to a continuous thioredoxin domain, and new termini that have been introduced in the helical domain by breaking the peptide bond Thr99-Gln100. cpDsbA(Q100T99) is catalytically active in vivo and in vitro. The crystal structure of oxidized cpDsbA(Q100T99), determined by molecular replacement at 2.4 A resolution, was found to be very similar to that of wild-type DsbA. The lower thermodynamic stability of cpDsbA(Q100T99) relative to DsbA is associated with small structural changes within the molecule, especially near the new termini and the circularizing linker. The active-site helices and adjacent loops display increased flexibility compared with oxidized DsbA.
Organizational Affiliation:
Forschungsgruppe Kristallographie, Max-Delbrück-Centrum für Molekulare Medizin, D-13092 Berlin, Germany.