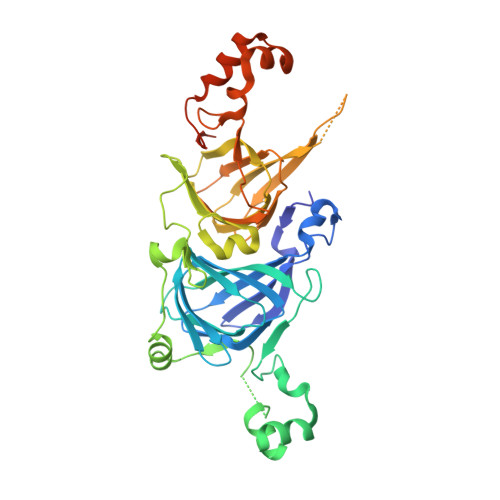
Structure of the core region of the soybean beta-conglycinin alpha' subunit.
Maruyama, Y., Maruyama, N., Mikami, B., Utsumi, S.(2004) Acta Crystallogr D Biol Crystallogr 60: 289-297
- PubMed: 14747705
- DOI: https://doi.org/10.1107/S0907444903027367
- Primary Citation of Related Structures:
1UIK - PubMed Abstract:
The crystal structure of the core region of the alpha' subunit (alpha(c')) of soybean beta-conglycinin has been determined at 2.3 A resolution. alpha(c') was superimposed on the known crystal structure of the beta-conglycinin beta subunit with a small root-mean square deviation of 0.77 A, which is consistent with the high sequence identity of 75.5% between alpha(c') and the beta subunit. It is known that the thermal stability of the beta subunit is higher than that of the alpha' subunit and that their thermal stabilities are conferred by highly homologous core regions. Comparisons of the three-dimensional structures and primary sequences between alpha(c') and the beta subunit suggest that five factors account for this difference between subunits as regards the difference in thermal stability: (i) the total cavity volume is larger in alpha(c'), (ii) the cluster of charged residues at the intermonomer interface is smaller in alpha(c') and alpha(c') lacks the intermonomer salt bridge of the beta subunit, (iii) the solvent-accessible surface is more hydrophobic in alpha(c'), (iv) there are fewer proline residues in alpha(c') and (v) a loop region between helix 3 and strand J' in alpha(c') is more flexible owing to the insertion of five additional residues. Although more hydrogen bonds were found in alpha(c'), this difference should be more than compensated for by the combined contributions of these other factors.
Organizational Affiliation:
Laboratory of Food Quality Design and Development, Graduate School of Agriculture, Kyoto University, Gokasho, Uji, Kyoto 611-0011, Japan.