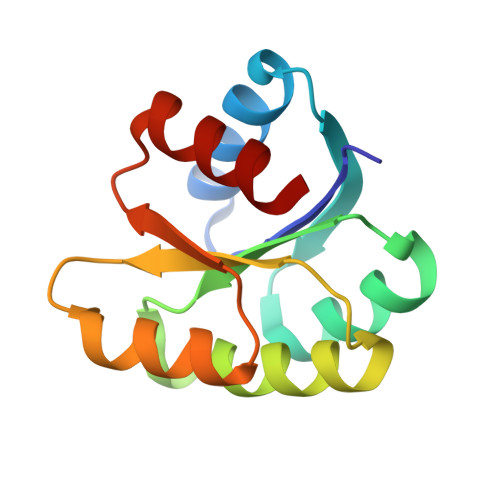
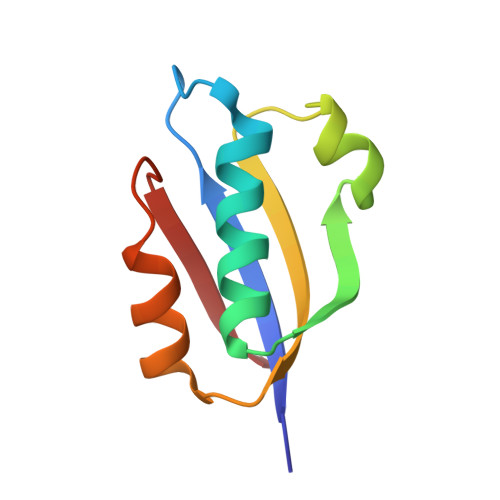
In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Park, S.Y., Beel, B.D., Simon, M.I., Bilwes, A.M., Crane, B.R.(2004) Proc Natl Acad Sci U S A 101: 11646-11651
- PubMed: 15289606
- DOI: https://doi.org/10.1073/pnas.0401038101
- Primary Citation of Related Structures:
1U0S - PubMed Abstract:
Although interfaces mediating protein-protein interactions are thought to be under strong evolutionary constraints, binding of the chemotaxis histidine kinase CheA to its phosphorylation target CheY suggests otherwise. The structure of Thermotoga maritima CheA domain P2 in complex with CheY reveals a different association than that observed for the same Escherichia coli proteins. Similar regions of CheY bind CheA P2 in the two systems, but the CheA P2 domains differ by an approximately 90 degrees rotation. CheA binds CheY with identical affinity in T. maritima and E. coli at the vastly different temperatures where the respective organisms live. Distinct sets of P2 residues mediate CheY binding in the two complexes; conservation patterns of these residues in CheA and compensations in CheY delineate two families of prokaryotic chemotaxis systems. A protein complex that has the same components and general function in different organisms, but an altered structure, indicates unanticipated complexity in the evolution of protein-protein interactions and cautions against extrapolating structural data from homologs.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14850, USA.