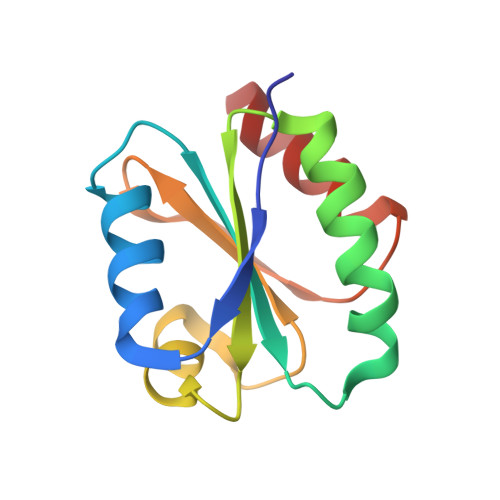
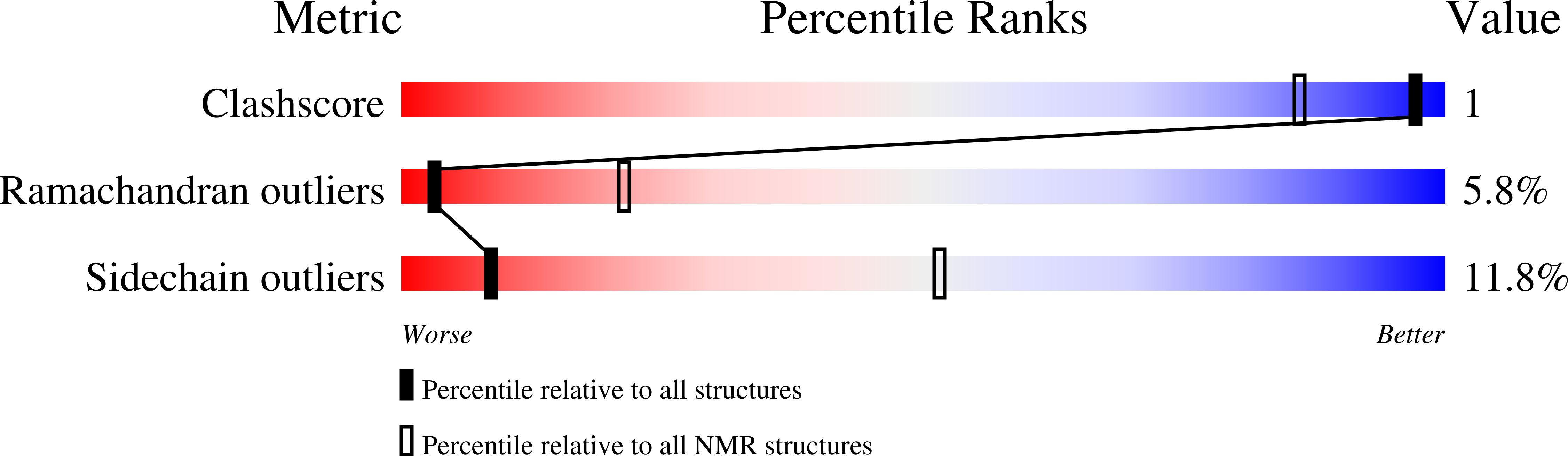Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar.
Coudevylle, N., Thureau, A., Hemmerlin, C., Gelhaye, E., Jacquot, J.P., Cung, M.T.(2005) Biochemistry 44: 2001-2008
- PubMed: 15697225
- DOI: https://doi.org/10.1021/bi047816n
- Primary Citation of Related Structures:
1TI3 - PubMed Abstract:
Assignment of heteronuclear and homonuclear multidimensional NMR spectra permits determination of the first three-dimensional solution structure of a higher-plant thioredoxin h. The collection of 1906 distance restraints, 137 TALOS-derived dihedral restraints, and 66 hydrogen bonds was used in the restrained molecular dynamics protocol to calculate the structure of the reduced form of thioredoxin h1 from poplar with an atomic rmsd of 0.60 +/- 0.12 A. This enzyme exhibits an unusual active site with the sequence WCPPC and original properties in terms of stability and specificity. Compared to other known thioredoxin structures, thioredoxin h1 from poplar adopts the classical "Trx fold". Its atypical active site possesses a conformation similar to that of other common thioredoxins but appears to be more rigid. Moreover, the hydrogen bond network, stabilizing the in-core beta-sheet, is tighter than in Chlamydomonas reinhardtii, explaining the difference in thermostability.
Organizational Affiliation:
Laboratoire de Chimie-Physique Macromoléculaire, UMR 7568 CNRS-INPL, Groupe ENSIC, 1 rue Grandville, B.P. 451, 54001 Nancy Cedex, France.