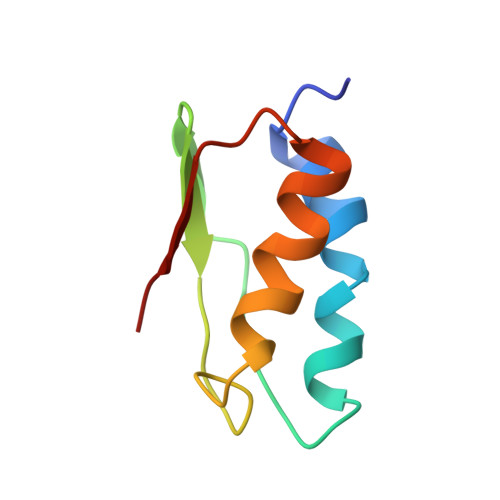
The NMR structure of the domain II of a chloroplastic NifU-like protein OsNifU1A.
Kumeta, H., Ogura, K., Asayama, M., Katoh, S., Katoh, E., Teshima, K., Inagaki, F.(2007) J Biomol NMR 38: 161-164
- PubMed: 17431550
- DOI: https://doi.org/10.1007/s10858-007-9155-9
- Primary Citation of Related Structures:
1TH5 - PubMed Abstract:
NifU-like proteins are a highly conserved protein that serves as the scaffold for assembly of Fe-S clusters. Chloroplastic NifU-like proteins have tandem NifU like domains, named domain I and domain II. Although the amino acid sequences of these domains are very similar to each other, the predicted functional region for the Fe-S cluster assembly, the CXXC motif, exists only in domain I. The structure of the domain II of chloroplastic NifU-like protein OsNifU1A has an alpha-beta sandwich structure containing two alpha helices located on one side of the beta-sheet. The electrostatic surface potential of OsNifU1A domain II is predominantly positively charged. Chloroplastic NifU-like proteins are targeted to ferredoxin for transferring the Fe-S cluster. The ferredoxin presents an overall negatively charged surface, which may evoke an electrostatic association with OsNifU1A domain II.
Organizational Affiliation:
Laboratory of Structural Biology, Graduate School of Pharmaceutical Sciences, Hokkaido University, Sapporo, Hokkaido, 060-0812, Japan.