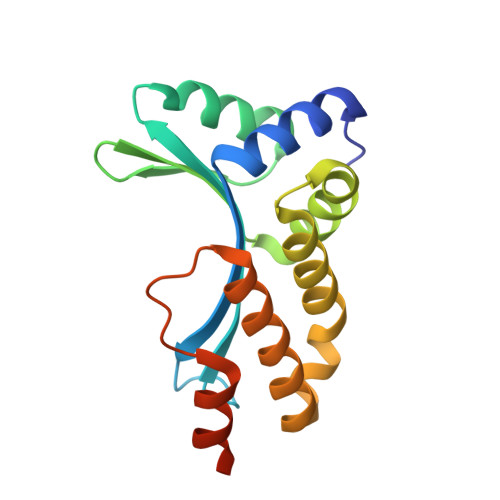
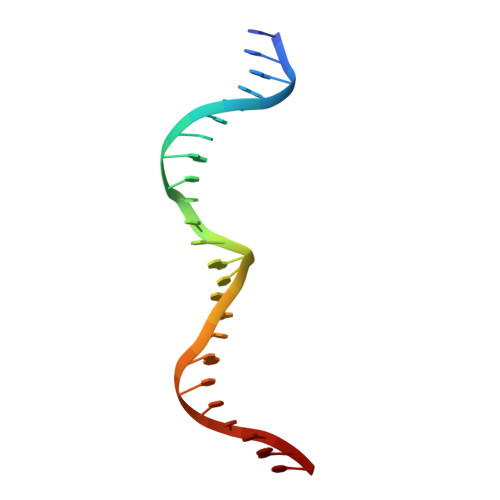
Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Chevalier, B., Sussman, D., Otis, C., Noel, A.J., Turmel, M., Lemieux, C., Stephens, K., Monnat Jr., R.J., Stoddard, B.L.(2004) Biochemistry 43: 14015-14026
- PubMed: 15518550
- DOI: https://doi.org/10.1021/bi048970c
- Primary Citation of Related Structures:
1T9I, 1T9J - PubMed Abstract:
The LAGLIDADG homing endonucleases include free-standing homodimers, pseudosymmetric monomers, and related enzyme domains embedded within inteins. DNA-bound structures of homodimeric I-CreI and monomeric I-SceI indicate that three catalytic divalent metal ions are distributed across a pair of overlapping active sites, with one shared metal participating in both strand cleavage reactions. These structures differ in the precise position and binding interactions of the metals. We have studied the metal dependence for the I-CreI homodimer using site-directed mutagenesis of active site residues and assays of binding affinity and cleavage activity. We have also reassessed the binding of a nonactivating metal ion (calcium) in the wild-type enzyme-substrate complex, and determined the DNA-bound structure of two inactive enzyme mutants. The conclusion of these studies is that the catalytic mechanism of symmetric LAGLIDADG homing endonucleases, and probably many of their monomeric cousins, involves a canonical two-metal mechanism in each of two active sites, which are chemically and structurally tethered to one another by a shared metal ion. Failure to occupy the shared metal site, as observed in the presence of calcium or when the metal-binding side chain from the LAGLIDADG motif (Asp 20) is mutated to asparagine, prevents cleavage by the enzyme.
Organizational Affiliation:
Graduate Program in Molecular and Cellular Biology, University of Washington and Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North A3-025, Seattle, Washington 98109, USA.