1T0K
Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex
- PDB DOI: https://doi.org/10.2210/pdb1T0K/pdb
- Entry: 1T0K supersedes: 1CK5, 1CK8, 1CN8, 1CN9
- NAKB: 1T0K
- Classification: RIBOSOME
- Organism(s): Escherichia coli, Saccharomyces cerevisiae
- Expression System: Escherichia coli
- Mutation(s): No
- Deposited: 2004-04-09 Released: 2004-07-20
Experimental Data Snapshot
- Method: X-RAY DIFFRACTION
- Resolution: 3.24 Å
- R-Value Free: 0.320
- R-Value Work: 0.261
- R-Value Observed: 0.269
wwPDB Validation 3D Report Full Report
This is version 2.1 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maltose-binding periplasmic protein | C [auth A] | 381 | Escherichia coli | Mutation(s): 0 Gene Names: MALE, B4034, Z5632, ECS5017 | 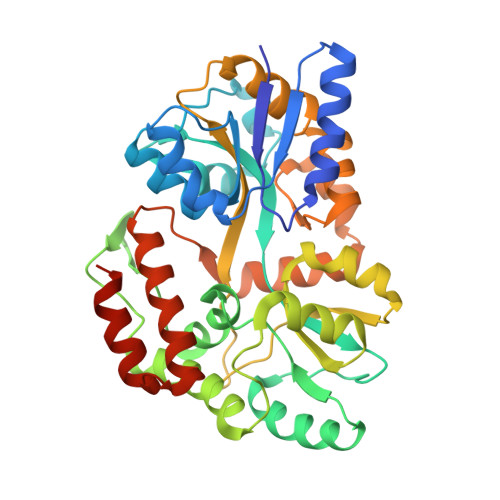 |
UniProt | |||||
Find proteins for P0AEX9 (Escherichia coli (strain K12)) Explore P0AEX9 Go to UniProtKB: P0AEX9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AEX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 60S ribosomal protein L30 | D [auth B] | 105 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: RPL30, RPL32, YGL030W | 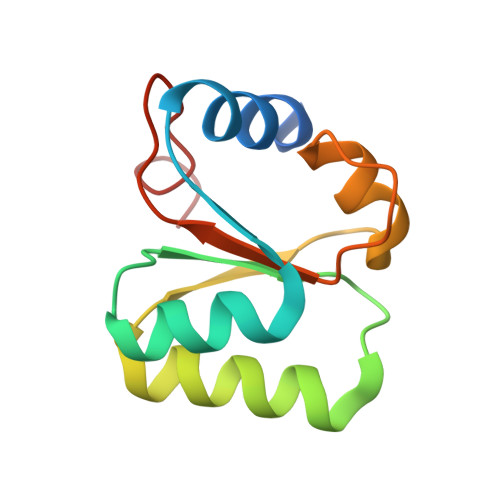 |
UniProt | |||||
Find proteins for P14120 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P14120 Go to UniProtKB: P14120 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14120 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3' | A [auth C] | 13 | N/A | 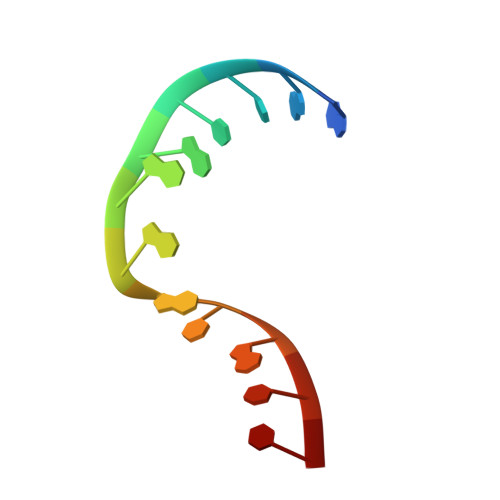 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3' | B [auth D] | 16 | N/A | 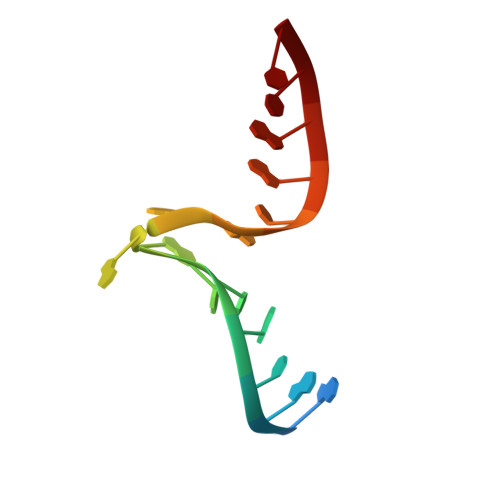 | |
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Biologically Interesting Molecules (External Reference) 1 Unique
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900010 Query on PRD_900010 | E | alpha-maltotetraose | Oligosaccharide / Substrate analog |  | |
Experimental Data & Validation
Experimental Data
- Method: X-RAY DIFFRACTION
- Resolution: 3.24 Å
- R-Value Free: 0.320
- R-Value Work: 0.261
- R-Value Observed: 0.269
- Space Group: P 41 21 2
Unit Cell:
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 136.006 | α = 90 |
| b = 136.006 | β = 90 |
| c = 123.813 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| AMoRE | phasing |
| CNS | refinement |
| CCP4 | data scaling |
Entry History
Deposition Data
- Released Date: 2004-07-20 Deposition Author(s): Chao, J.A., Williamson, J.R.
- This entry supersedes: 1CK5, 1CK8, 1CN8, 1CN9
Revision History (Full details and data files)
- Version 1.0: 2004-07-20
Type: Initial release - Version 1.1: 2008-04-30
Changes: Version format compliance - Version 1.2: 2011-07-13
Changes: Version format compliance - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Atomic model, Data collection, Database references, Derived calculations, Non-polymer description, Structure summary - Version 2.1: 2022-12-21
Changes: Database references, Structure summary














