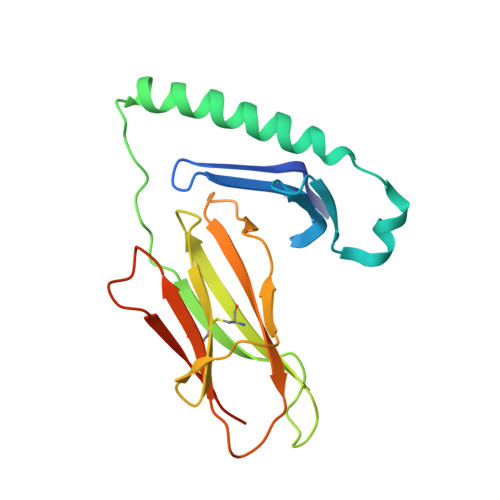
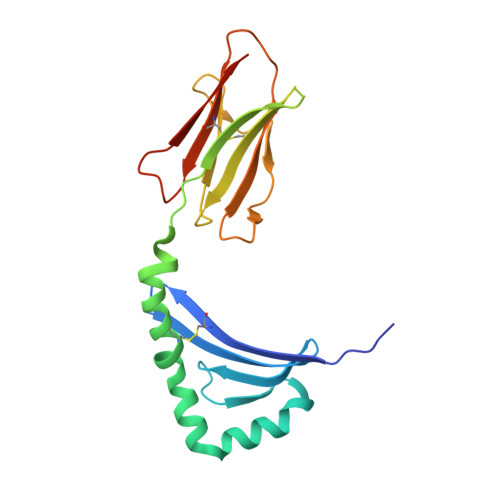
Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease
Kim, C.-Y., Quarsten, H., Bergseng, E., Khosla, C., Sollid, L.M.(2004) Proc Natl Acad Sci U S A 101: 4175-4179
- PubMed: 15020763
- DOI: https://doi.org/10.1073/pnas.0306885101
- Primary Citation of Related Structures:
1S9V - PubMed Abstract:
Celiac disease, also known as celiac sprue, is a gluten-induced autoimmune-like disorder of the small intestine, which is strongly associated with HLA-DQ2. The structure of DQ2 complexed with an immunogenic epitope from gluten, QLQPFPQPELPY, has been determined to 2.2-A resolution by x-ray crystallography. The glutamate at P6, which is formed by tissue transglutaminase-catalyzed deamidation, is an important anchor residue as it participates in an extensive hydrogen-bonding network involving Lys-beta71 of DQ2. The gluten peptide-DQ2 complex retains critical hydrogen bonds between the MHC and the peptide backbone despite the presence of many proline residues in the peptide that are unable to participate in amide-mediated hydrogen bonds. Positioning of proline residues such that they do not interfere with backbone hydrogen bonding results in a reduction in the number of registers available for gluten peptides to bind to MHC class II molecules and presumably impairs the likelihood of establishing favorable side-chain interactions. The HLA association in celiac disease can be explained by a superior ability of DQ2 to bind the biased repertoire of proline-rich gluten peptides that have survived gastrointestinal digestion and that have been deamidated by tissue transglutaminase. Finally, surface-exposed proline residues in the proteolytically resistant ligand were replaced with functionalized analogs, thereby providing a starting point for the design of orally active agents for blocking gluten-induced toxicity.
Organizational Affiliation:
Department of Chemical Engineering, Stanford University, Stanford, CA 94305, USA.