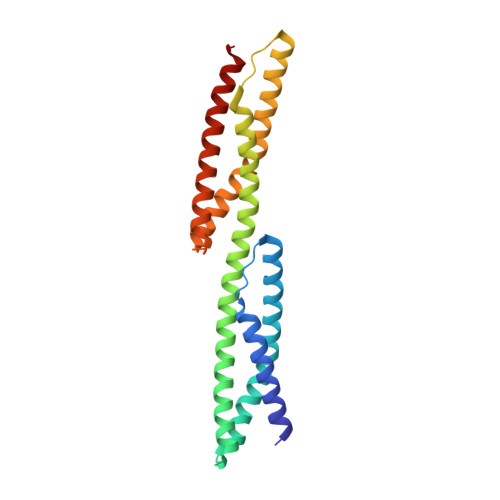
Structural insights into the stability and flexibility of unusual erythroid spectrin repeats
Kusunoki, H., MacDonald, R.I., Mondragon, A.(2004) Structure 12: 645-656
- PubMed: 15062087
- DOI: https://doi.org/10.1016/j.str.2004.02.022
- Primary Citation of Related Structures:
1S35 - PubMed Abstract:
Erythroid spectrin, a major component of the cytoskeletal network of the red cell which contributes to both the stability and the elasticity of the red cell membrane, is composed of two subunits, alpha and beta, each formed by 16-20 tandem repeats. The properties of the repeats and their relative arrangement are thought to be key determinants of spectrin flexibility. Here we report a 2.4 A resolution crystal structure of human erythroid beta-spectrin repeats 8 and 9. This two-repeat fragment is unusual as it exhibits low stability of folding and one of its repeats lacks two tryptophans highly conserved among spectrin repeats. Two key factors responsible for the lower stability and, possibly, its flexibility, are revealed by the structure. A third novel feature of the structure is the relative orientation of the two repeats, which increases the range of possible conformations and provides new insights into atomic models of spectrin flexibility.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, 2205 Tech Drive, Evanston, IL 60208 USA.