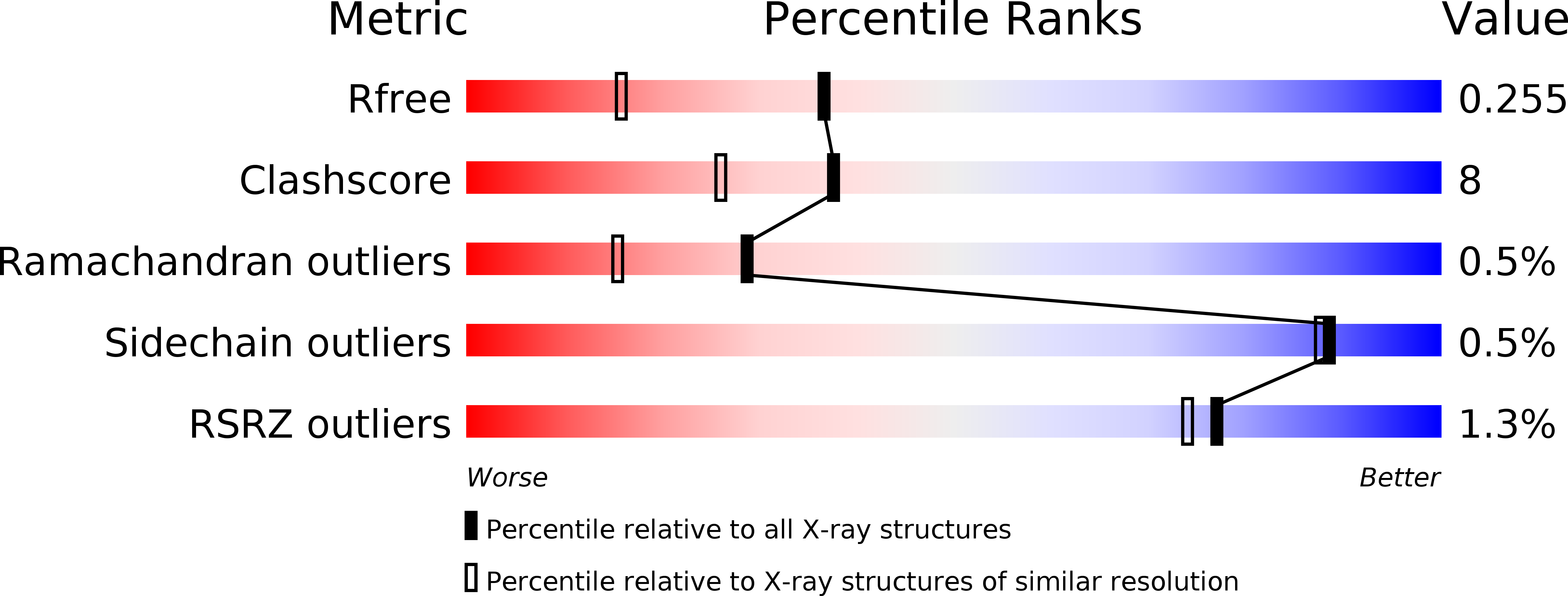
Trimeric structure of a C-type mannose-binding protein.
Weis, W.I., Drickamer, K.(1994) Structure 2: 1227-1240
- PubMed: 7704532
- DOI: https://doi.org/10.1016/S0969-2126(94)00124-3
- Primary Citation of Related Structures:
1RTM - PubMed Abstract:
Mannose-binding proteins (MBPs) are C-type (Ca(2+)-dependent) animal lectins found in serum. They recognize cell-surface oligosaccharide structures characteristic of pathogenic bacteria and fungi, and trigger the neutralization of these organisms. Like most lectins, MBPs display weak intrinsic affinity for monovalent sugar ligands, but bind avidly to multivalent ligands.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, CA 94305.