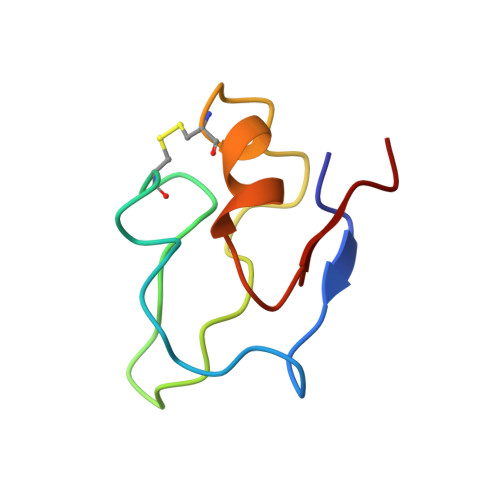
An NMR-derived model for the solution structure of oxidized Thermotoga maritima 1[Fe4-S4] ferredoxin.
Sticht, H., Wildegger, G., Bentrop, D., Darimont, B., Sterner, R., Rosch, P.(1996) Eur J Biochem 237: 726-735
- PubMed: 8647119
- DOI: https://doi.org/10.1111/j.1432-1033.1996.0726p.x
- Primary Citation of Related Structures:
1ROF - PubMed Abstract:
The solution structure of the 60-residue 1[Fe4-S4] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima was determined based on 683 distance and 35 dihedral angle restraints that were obtained from NMR data. In addition, data known from crystallographic studies of ferredoxins was used for modeling of the iron-sulfur cluster and its environment. The protein shows a globular fold very similar to the fold of the related 1[Fe4-S4] ferredoxins from Desulfovibrio gigas and Desulfovibrio africanus, and elements of regular secondary structure similar to those in other ferredoxins were found in the T. maritima protein. In particular, the T. maritima protein displayed a beta-sheet structure made up of strands located at the very NH(2) and COOH termini of the protein, and an internal alpha-helix. The internal beta-sheet observed in the D. gigas and D. africanus ferredoxins could not be confirmed in T. maritima ferredoxin and is thus suggested to be only weakly present or even absent in this protein. This result suggests that thermostability in ferredoxins is not necessarily correlated with the content of stable elements of regular secondary structure.
Organizational Affiliation:
Lehrstuhl für Biopolymere, Universität Bayreuth, Germany.