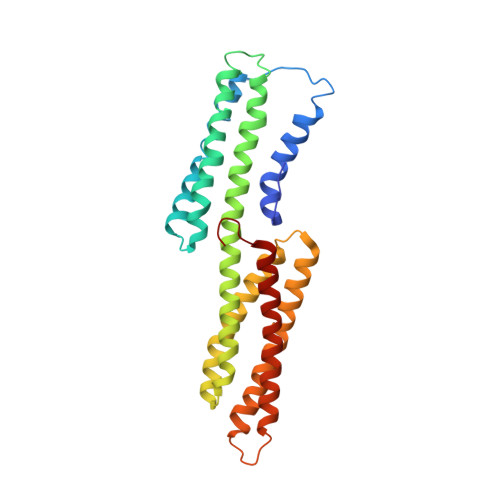
Vinculin activation by talin through helical bundle conversion
Izard, T., Evans, G., Borgon, R.A., Rush, C.L., Bricogne, G., Bois, P.R.(2004) Nature 427: 171-175
- PubMed: 14702644
- DOI: https://doi.org/10.1038/nature02281
- Primary Citation of Related Structures:
1RKC, 1RKE - PubMed Abstract:
Vinculin is a conserved component and an essential regulator of both cell-cell (cadherin-mediated) and cell-matrix (integrin-talin-mediated focal adhesions) junctions, and it anchors these adhesion complexes to the actin cytoskeleton by binding to talin in integrin complexes or to alpha-actinin in cadherin junctions. In its resting state, vinculin is held in a closed conformation through interactions between its head (Vh) and tail (Vt) domains. The binding of vinculin to focal adhesions requires its association with talin. Here we report the crystal structures of human vinculin in its inactive and talin-activated states. Talin binding induces marked conformational changes in Vh, creating a novel helical bundle structure, and this alteration actively displaces Vt from Vh. These results, as well as the ability of alpha-actinin to also bind to Vh and displace Vt from pre-existing Vh-Vt complexes, support a model whereby Vh functions as a domain that undergoes marked structural changes that allow vinculin to direct cytoskeletal assembly in focal adhesions and adherens junctions. Notably, talin's effects on Vh structure establish helical bundle conversion as a signalling mechanism by which proteins direct cellular responses.
Organizational Affiliation:
Department of Hematology-Oncology, St Jude Children's Research Hospital, Memphis, Tennessee 38105, USA. tina.izard@stjude.org