The crystal structure of Rv2991 from Mycobacterium tuberculosis: An F420binding protein with unknown function.
Benini, S., Haouz, A., Proux, F., Alzari, P., Wilson, K.(2019) J Struct Biol
- PubMed: 30890426
- DOI: https://doi.org/10.1016/j.jsb.2019.03.006
- Primary Citation of Related Structures:
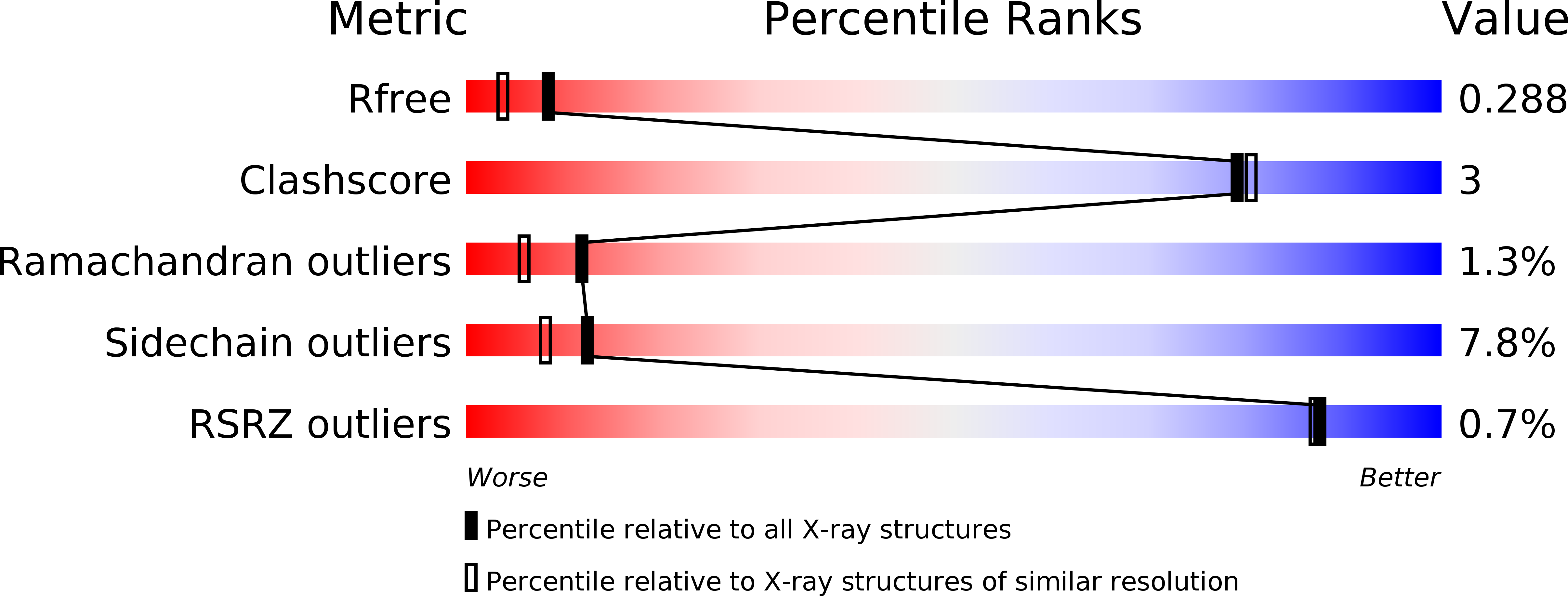
1RFE - PubMed Abstract:
The crystal structure of the conserved hypothetical protein Rv2991 from Mycobacterium tuberculosis has been solved by SAD using seleno-methionine substituted protein. The dimeric biological assembly and the sequence and fold conservation are typical of F 420 cofactor binding enzymes. Despite Rv2991 still being of unknown function, sequence and structural comparison with similar proteins enable a role to be proposed for its C-terminal stretch of residues in recognizing and orienting the substrate. In addition, the C-terminus is involved in both protein folding and determining the size of the active site cavity.
Organizational Affiliation:
Bioorganic Chemistry and Bio-Crystallography Laboratory (B(2)Cl), Faculty of Science and Technology, Free University of Bolzano, Piazza Università 5, Bolzano 39100, Italy. Electronic address: stefano.benini@unibz.it.