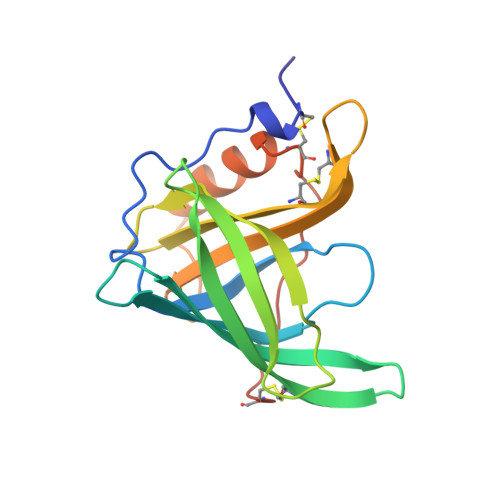
Crystallographic refinement of human serum retinol binding protein at 2A resolution.
Cowan, S.W., Newcomer, M.E., Jones, T.A.(1990) Proteins 8: 44-61
- PubMed: 2217163
- DOI: https://doi.org/10.1002/prot.340080108
- Primary Citation of Related Structures:
1RBP - PubMed Abstract:
Human serum retinol binding protein (RBP) in complex with retinol has been crystallographically refined to an R-factor of 18.1% with 2A resolution data. The protein topology results in an anti-parallel beta-barrel that encapsulates the retinol ligand. A detailed description of the protein and the binding site is provided. Our structural work has helped to define a family of proteins, many of which are carrier proteins for smaller ligand molecules. We describe the structural basis for the conservation of sequence within the family.
Organizational Affiliation:
Department of Molecular Biology, Biomedicum Centre, Uppsala, Sweden.