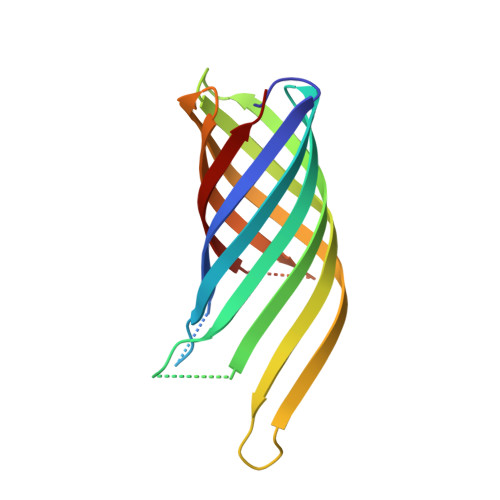
High Resolution Structure of the Ompa Membrane Domain
Pautsch, A., Schulz, G.E.(2000) J Mol Biol 298: 273
- PubMed: 10764596
- DOI: https://doi.org/10.1006/jmbi.2000.3671
- Primary Citation of Related Structures:
1QJP - PubMed Abstract:
The membrane domain of OmpA consists of an eight-stranded all-next-neighbor antiparallel beta-barrel with short turns at the periplasmic barrel end and long flexible loops at the external end. The structure analysis has been extended from medium resolution to 1. 65 A (1 A=0.1 nm), and the molecular model has been refined anisotropically to show oriented mobilities of the structural elements. The improved data allowed us to locate five further detergent molecules and 11 more water molecules. Moreover, the two large non-polar packing contacts have now been defined in detail. The analysis indicates that the beta-barrel constitutes a solid scaffold such that the long external loops need not contribute to stability. These loops are highly mobile and thus cause a major problem during the crystallization process. The beta-barrel was related to those of lipocalins. Two further crystal forms with exceptionally dense packing arrangements were established at medium resolution.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität, Albertstrasse 21, Freiburg im Breisgau, D-79104, Germany.