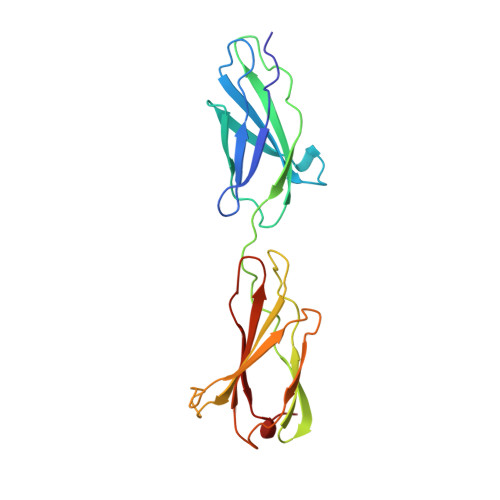
Crystal structure of a tandem pair of fibronectin type III domains from the cytoplasmic tail of integrin alpha6beta4.
de Pereda, J.M., Wiche, G., Liddington, R.C.(1999) EMBO J 18: 4087-4095
- PubMed: 10428948
- DOI: https://doi.org/10.1093/emboj/18.15.4087
- Primary Citation of Related Structures:
1QG3 - PubMed Abstract:
The integrin alpha6beta4 is an essential component of hemidesmosomes but it also plays a dynamic role in invasive carcinoma cells. The cytoplasmic tail of the beta4 subunit is uniquely large among integrins and includes two pairs of fibronectin type III domains separated by a connecting segment. Here we describe the crystal structure of the first tandem domain pair, a module that is critical for alpha6beta4 function. The structure reveals a novel interdomain interface and candidate protein-binding sites, including a large acidic cleft formed from the surfaces of both domains and a prominent loop that is reminiscent of the RGD integrin-binding loop of fibronectin. This is the first crystal structure of either a hemidesmosome component or an integrin cytoplasmic domain, and it will enable the intracellular functions of alpha6beta4 to be dissected at the atomic level.
Organizational Affiliation:
Department of Biochemistry, University of Leicester, Leicester LE1 7RH, UK.