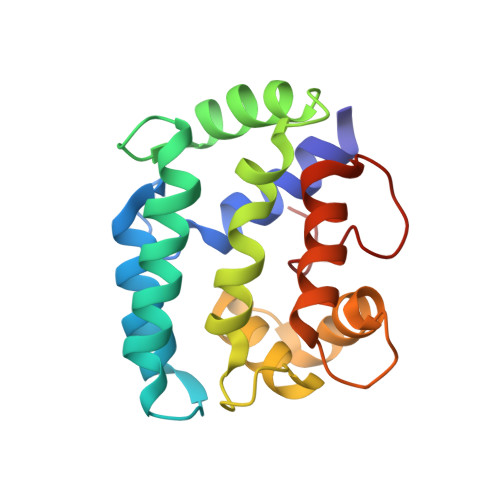
Solution structure and internal dynamics of NSCP, a compact calcium-binding protein.
Rabah, G., Popescu, R., Cox, J.A., Engelborghs, Y., Craescu, C.T.(2005) FEBS J 272: 2022-2036
- PubMed: 15819893
- DOI: https://doi.org/10.1111/j.1742-4658.2005.04629.x
- Primary Citation of Related Structures:
1Q80 - PubMed Abstract:
The solution structure of Nereis diversicolor sarcoplasmic calcium-binding protein (NSCP) in the calcium-bound form was determined by NMR spectroscopy, distance geometry and simulated annealing. Based on 1859 NOE restraints and 262 angular restraints, 17 structures were generated with a rmsd of 0.87 A from the mean structure. The solution structure, which is highly similar to the structure obtained by X-ray crystallography, includes two open EF-hand domains, which are in close contact through their hydrophobic surfaces. The internal dynamics of the protein backbone were determined by studying amide hydrogen/deuterium exchange rates and 15N nuclear relaxation. The two methods revealed a highly compact and rigid structure, with greatly restricted mobility at the two termini. For most of the amide protons, the free energy of exchange-compatible structural opening is similar to the free energy of structural stability, suggesting that isotope exchange of these protons takes place through global unfolding of the protein. Enhanced conformational flexibility was noted in the unoccupied Ca2+-binding site II, as well as the neighbouring helices. Analysis of the experimental nuclear relaxation and the molecular dynamics simulations give very similar profiles for the backbone generalized order parameter (S2), a parameter related to the amplitude of fast (picosecond to nanosecond) movements of N(H)-H vectors. We also noted a significant correlation between this parameter, the exchange rate, and the crystallographic B factor along the sequence.
Organizational Affiliation:
INSERM & Institut Curie, Centre Universitaire, Orsay, France.