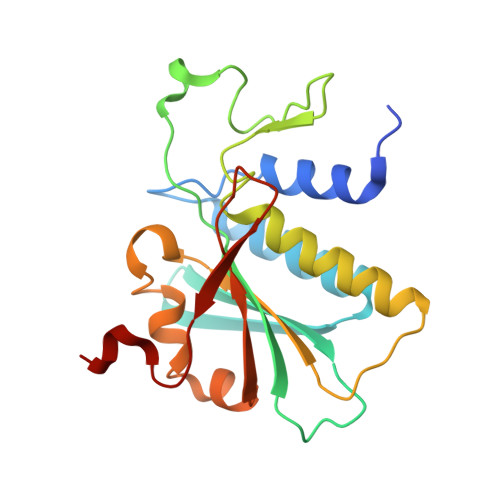
Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family.
Lindner, H.A., Lunin, V.V., Alary, A., Hecker, R., Cygler, M., Menard, R.(2003) J Biol Chem 278: 44496-44504
- PubMed: 12933810
- DOI: https://doi.org/10.1074/jbc.M304233200
- Primary Citation of Related Structures:
1Q7L - PubMed Abstract:
Members of the aminoacylase-1 (Acy1)/M20 family of aminoacylases and exopeptidases exist as either monomers or homodimers. They contain a zinc-binding domain and a second domain mediating dimerization in the latter case. The roles that both domains play in catalysis have been investigated for human Acy1 (hAcy1) by x-ray crystallography and by site-directed mutagenesis. Structure comparison of the dinuclear zinc center in a mutant of hAcy1 reported here with dizinc centers in related enzymes points to a difference in zinc ligation in the Acy1/M20 family. Mutational analysis supports catalytic roles of zinc ions, a vicinal glutamate, and a histidine from the dimerization domain. By complementing different active site mutants of hAcy1, we show that catalysis occurs at the dimer interface. Reinterpretation of the structure of a monomeric homolog, peptidase V, reveals that a domain insertion mimics dimerization. We conclude that monomeric and dimeric Acy1/M20 family members share a unique active site architecture involving both enzyme domains. The study may provide means to improve homologous carboxypeptidase G2 toward application in antibody-directed enzyme prodrug therapy.
Organizational Affiliation:
Biotechnology Research Institute, National Research Council of Canada, Montréal, Québec H4P 2R2, Canada.