The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex.
Cheng, K.Y., Lowe, E.D., Sinclair, J., Nigg, E.A., Johnson, L.N.(2003) EMBO J 22: 5757-5768
- PubMed: 14592974
- DOI: https://doi.org/10.1093/emboj/cdg558
- Primary Citation of Related Structures:
1Q4K, 1Q4O - PubMed Abstract:
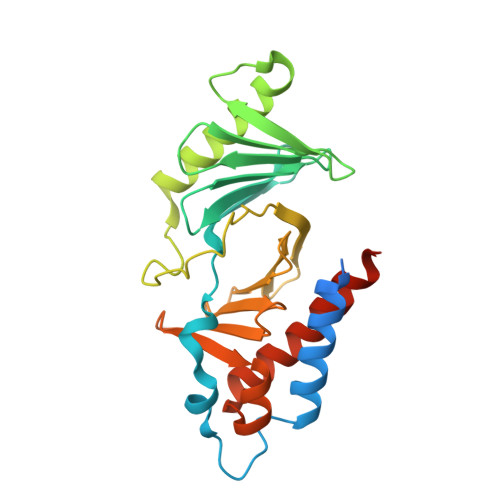
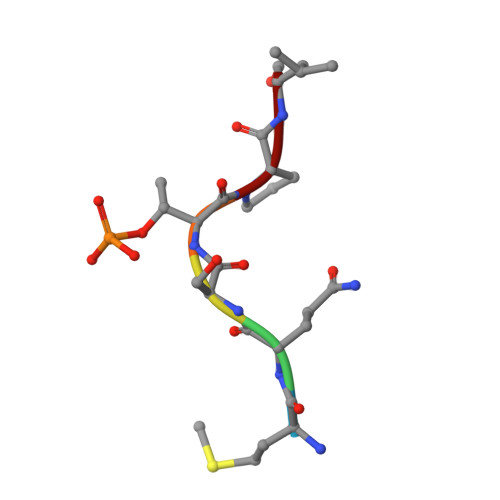
Human polo-like kinase Plk1 localizes to the centrosomes, kinetochores and central spindle structures during mitosis. It plays an essential role in promoting mitosis and cytokinesis through phosphorylation of a number of different substrates. Kinase activity is regulated by a conserved C-terminal domain, termed the polo box domain (PBD), which acts both as an autoinhibitory domain and as a subcellular localization domain. We have determined the crystal structure of Plk1 PBD (residues 367-603) to 2.2 A resolution and the structure of a phospho-peptide-PBD (residues 345-603) complex to 2.3 A resolution. The two polo boxes of the PBD exhibit identical folds based on a six-stranded beta-sheet and an alpha-helix, despite only 12% sequence identity. The phospho-peptide binds at a site between the two polo boxes. It makes a short antiparallel beta-sheet connection and critical contacts to residues Trp414, Leu490, His538 and Lys540. Most of these residues had been shown to be important for biological activity through mutational studies. The results provide an explanation for phospho-peptide recognition and create the basis for new functional studies.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.