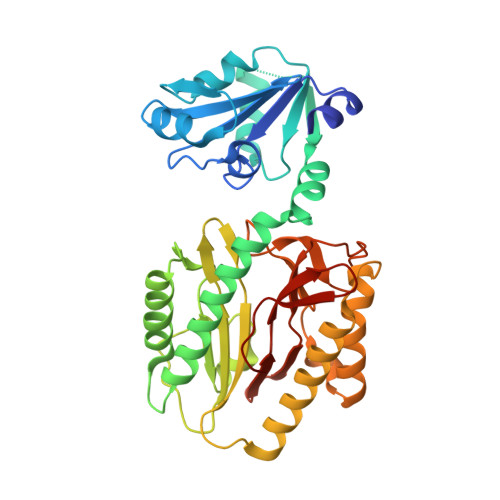
Structure of the Prolidase from Pyrococcus furiosus.
Maher, M.J., Ghosh, M., Grunden, A.M., Menon, A.L., Adams, M.W., Freeman, H.C., Guss, J.M.(2004) Biochemistry 43: 2771-2783
- PubMed: 15005612
- DOI: https://doi.org/10.1021/bi0356451
- Primary Citation of Related Structures:
1PV9 - PubMed Abstract:
The structure of prolidase from the hyperthermophilic archaeon Pyrococcus furiosus (Pfprol) has been solved and refined at 2.0 A resolution. This is the first structure of a prolidase, i.e., a peptidase specific for dipeptides having proline as the second residue. The asymmetric unit of the crystals contains a homodimer of the enzyme. Each of the two protein subunits has two domains. The C-terminal domain includes the catalytic site, which is centered on a dinuclear metal cluster. In the as-isolated form of Pfprol, the active-site metal atoms are Co(II) [Ghosh, M., et al. (1998) J. Bacteriol. 180, 4781-9]. An unexpected finding is that in the crystalline enzyme the active-site metal atoms are Zn(II), presumably as a result of metal exchange during crystallization. Both of the Zn(II) atoms are five-coordinate. The ligands include a bridging water molecule or hydroxide ion, which is likely to act as a nucleophile in the catalytic reaction. The two-domain polypeptide fold of Pfprol is similar to the folds of two functionally related enzymes, aminopeptidase P (APPro) and creatinase. In addition, the catalytic C-terminal domain of Pfprol has a polypeptide fold resembling that of the sole domain of a fourth enzyme, methionine aminopeptidase (MetAP). The active sites of APPro and MetAP, like that of Pfprol, include a dinuclear metal center. The metal ligands in the three enzymes are homologous. Comparisons with the molecular structures of APPro and MetAP suggest how Pfprol discriminates against oligopeptides and in favor of Xaa-Pro substrates. The crystal structure of Pfprol was solved by multiple-wavelength anomalous dispersion. The crystals yielded diffraction data of relatively high quality and resolution, despite the fact that one of the two protein subunits in the asymmetric unit was found to be significantly disordered. The final R and R(free) values are 0.24 and 0.28, respectively.
Organizational Affiliation:
School of Molecular and Microbial Biosciences, University of Sydney, New South Wales 2006, Australia.