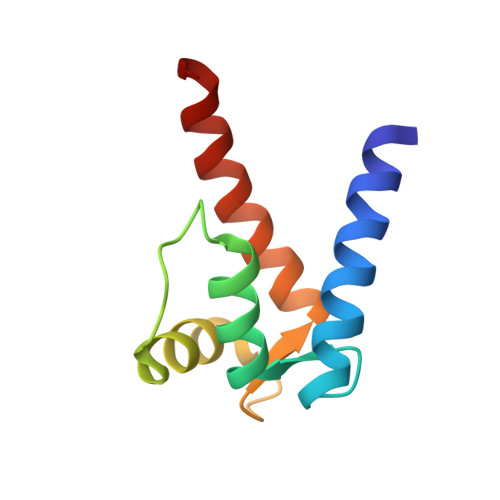
Structure of the Ca(2+)/S100B/NDR Kinase Peptide Complex: Insights into S100 Target Specificity and Activation of the Kinase.
Bhattacharya, S., Large, E., Heizmann, C.W., Hemmings, B., Chazin, W.J.(2003) Biochemistry 42: 14416-14426
- PubMed: 14661952
- DOI: https://doi.org/10.1021/bi035089a
- Primary Citation of Related Structures:
1PSB - PubMed Abstract:
NDR, a nuclear serine/threonine kinase, belongs to the subfamily of Dbf2 kinases that is critical to the morphology and proliferation of cells. The activity of NDR kinase is modulated in a Ca(2+)/S100B-dependent manner by phosphorylation of Ser281 in the catalytic domain and Thr444 in the C-terminal regulatory domain. S100B, which is a member of the S100 subfamily of EF-hand proteins, binds to a basic/hydrophobic sequence at the junction of the N-terminal regulatory and catalytic domains (NDR(62-87)). Unlike calmodulin-dependent kinases, regulation of NDR by S100B is not associated with direct autoinhibition of the active site, but rather involves a conformational change in the catalytic domain triggered by Ca(2+)/S100B binding to the junction region. To gain further insight into the mechanism of activation of the kinase, studies have been carried out on Ca(2+)/S100B in complex with the intact N-terminal regulatory domain, NDR(1-87). Multidimensional heteronuclear NMR analysis showed that the binding mode and stoichiometry of a peptide fragment of NDR (NDR(62-87)) is the same as for the intact N-terminal regulatory domain. The solution structure of Ca(2+)/S100B and NDR(62-87) has been determined. One target molecule is found to associate with each subunit of the S100B dimer. The peptide adopts three turns of helix in the bound state, and the complex is stabilized by both hydrophobic and electrostatic interactions. These structural studies, in combination with available biochemical data, have been used to develop a model for calcium-induced activation of NDR kinase by S100B.
Organizational Affiliation:
Department of Biochemistry, Center for Structural Biology, Vanderbilt University, Nashville, Tennessee 37232-8725, USA.